Please be patient as the page loads
|
SORC3_MOUSE
|
||||||
| SwissProt Accessions | Q8VI51, Q9CTR4 | Gene names | Sorcs3 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SORC1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.993316 (rank : 3) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WY21, Q59GG7, Q5JVT7, Q5JVT8, Q5VY14, Q86WQ1, Q86WQ2, Q9H1Y1, Q9H1Y2 | Gene names | SORCS1, SORCS | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS1 precursor (hSorCS). | |||||
|
SORC1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.992814 (rank : 4) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JLC4, Q8VI45, Q922I1, Q9QY21 | Gene names | Sorcs1, Sorcs | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS1 precursor (mSorCS). | |||||
|
SORC2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.978401 (rank : 6) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96PQ0, Q9P2L7 | Gene names | SORCS2, KIAA1329 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS2 precursor. | |||||
|
SORC2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.982754 (rank : 5) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EPR5 | Gene names | Sorcs2 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS2 precursor. | |||||
|
SORC3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.994879 (rank : 2) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPU3, Q5VXF9, Q9NQJ2 | Gene names | SORCS3, KIAA1059 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
SORC3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8VI51, Q9CTR4 | Gene names | Sorcs3 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
SORL_MOUSE
|
||||||
| θ value | 1.5555e-68 (rank : 7) | NC score | 0.458969 (rank : 9) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88307, O54711, O70581 | Gene names | Sorl1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (mSorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11) (Gp250). | |||||
|
SORL_HUMAN
|
||||||
| θ value | 7.71989e-68 (rank : 8) | NC score | 0.455247 (rank : 10) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92673, Q92856 | Gene names | SORL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (SorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11). | |||||
|
SORT_HUMAN
|
||||||
| θ value | 7.49467e-55 (rank : 9) | NC score | 0.858637 (rank : 8) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99523, Q8IZ49 | Gene names | SORT1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin precursor (Neurotensin receptor 3) (NTR3) (NT3) (Glycoprotein 95) (Gp95) (100 kDa NT receptor). | |||||
|
SORT_MOUSE
|
||||||
| θ value | 8.28633e-54 (rank : 10) | NC score | 0.859378 (rank : 7) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PHU5, Q3UHE2, Q8K043, Q9QXW6 | Gene names | Sort1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sortilin precursor (Neurotensin receptor 3) (NTR3) (mNTR3). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 11) | NC score | 0.021811 (rank : 27) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
ANR28_MOUSE
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.012130 (rank : 42) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q505D1 | Gene names | Ankrd28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR28_HUMAN
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.011843 (rank : 43) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15084, Q6ULS0, Q6ZT57 | Gene names | ANKRD28, KIAA0379 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
CUL7_HUMAN
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.033024 (rank : 25) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14999, Q5T654 | Gene names | CUL7, KIAA0076 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-7 (CUL-7). | |||||
|
T22D4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.025120 (rank : 26) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
GCH1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.048251 (rank : 24) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P30793, Q9Y4I8 | Gene names | GCH1, DYT5, GCH | |||
|
Domain Architecture |

|
|||||
| Description | GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I). | |||||
|
GPR61_HUMAN
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.014551 (rank : 36) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZJ8, Q6NWS0, Q8TDV4, Q96PR4 | Gene names | GPR61, BALGR | |||
|
Domain Architecture |
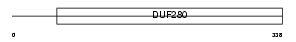
|
|||||
| Description | Probable G-protein coupled receptor 61 (Biogenic amine receptor-like G-protein coupled receptor). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.007490 (rank : 49) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PKD1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.049884 (rank : 23) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.015968 (rank : 34) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
GPR61_MOUSE
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.012605 (rank : 41) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 702 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C010 | Gene names | Gpr61 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 61. | |||||
|
CDYL1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.011008 (rank : 45) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y232 | Gene names | CDYL, CDYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
PHLD2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.017541 (rank : 32) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15127 | Gene names | GPLD2, PIGPLD2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-glycan-specific phospholipase D 2 precursor (EC 3.1.4.50) (PI-G PLD) (Glycoprotein phospholipase D) (Glycosyl- phosphatidylinositol-specific phospholipase D). | |||||
|
AOC3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.019245 (rank : 31) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16853, Q45F94 | Gene names | AOC3, VAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane copper amine oxidase (EC 1.4.3.6) (Semicarbazide-sensitive amine oxidase) (SSAO) (Vascular adhesion protein 1) (VAP-1) (HPAO). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.009536 (rank : 47) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
PAPPA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.020709 (rank : 29) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13219, Q08371, Q68G52, Q9UDK7 | Gene names | PAPPA | |||
|
Domain Architecture |

|
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
PKHA4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.012859 (rank : 40) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
WEE1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.001506 (rank : 52) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 794 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47810, Q9EPL7 | Gene names | Wee1 | |||
|
Domain Architecture |

|
|||||
| Description | Wee1-like protein kinase (EC 2.7.10.2) (Wee1A kinase). | |||||
|
ZN206_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | -0.001295 (rank : 57) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96SZ4 | Gene names | ZNF206, ZSCAN10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 206 (Zinc finger and SCAN domain-containing protein 10). | |||||
|
ZN541_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.013505 (rank : 39) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.013763 (rank : 38) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.020638 (rank : 30) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
MINK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | -0.000872 (rank : 55) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1451 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JM52, Q921M6, Q9JM92 | Gene names | Mink1, Map4k6, Mink | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.008242 (rank : 48) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
PK1L1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.020830 (rank : 28) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
PRKDC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.011322 (rank : 44) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97313, O88187, P97928, Q307W9, Q3V2W8, Q8C2A7, Q9Z341 | Gene names | Prkdc, Xrcc7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (P460). | |||||
|
ZN219_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.000451 (rank : 53) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 876 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P2Y4, Q8IYC1, Q9BW28 | Gene names | ZNF219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 219. | |||||
|
GPNMB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.016770 (rank : 33) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14956, Q6UVX1, Q8N1A1 | Gene names | GPNMB, HGFIN, NMB | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane glycoprotein NMB precursor (Transmembrane glycoprotein HGFIN). | |||||
|
NLGN2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.003350 (rank : 51) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q69ZK9, Q5F288 | Gene names | Nlgn2, Kiaa1366 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroligin-2 precursor. | |||||
|
AATK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | -0.000649 (rank : 54) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
CT012_HUMAN
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.014409 (rank : 37) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NVP4, Q4F7X1, Q5QPD9, Q5QPE0, Q68DN8, Q6ZMX9, Q96NF0, Q9H1E0, Q9H442 | Gene names | C20orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C20orf12. | |||||
|
FUCO2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.006291 (rank : 50) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BTY2, Q7Z6Y2, Q8NBK4 | Gene names | FUCA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plasma alpha-L-fucosidase precursor (EC 3.2.1.51) (Alpha-L-fucosidase 2) (Alpha-L-fucoside fucohydrolase 2). | |||||
|
MINK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | -0.001101 (rank : 56) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N4C8, Q9P1X1, Q9P2R8 | Gene names | MINK1, MAP4K6, MINK | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
PHLD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.010717 (rank : 46) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P80108, Q15128 | Gene names | GPLD1, PIGPLD1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-glycan-specific phospholipase D 1 precursor (EC 3.1.4.50) (PI-G PLD) (Glycoprotein phospholipase D) (Glycosyl- phosphatidylinositol-specific phospholipase D). | |||||
|
PME17_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.014740 (rank : 35) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60696 | Gene names | Silv, D10H12S53E, Pmel17, Si | |||
|
Domain Architecture |

|
|||||
| Description | Melanocyte protein Pmel 17 precursor (Silver locus protein). | |||||
|
CD320_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.065857 (rank : 11) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NPF0, Q53HF7 | Gene names | CD320, 8D6A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD320 antigen precursor (8D6 antigen) (FDC-signaling molecule 8D6) (FDC-SM-8D6). | |||||
|
LDLR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.050030 (rank : 22) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35951 | Gene names | Ldlr | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor precursor (LDL receptor). | |||||
|
LRP12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.050697 (rank : 16) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BUJ9, Q8BWM9 | Gene names | Lrp12 | |||
|
Domain Architecture |
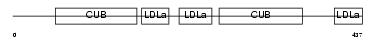
|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor. | |||||
|
LRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.050398 (rank : 19) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
LRP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.054822 (rank : 12) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
LRP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.051018 (rank : 15) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75074 | Gene names | LRP3 | |||
|
Domain Architecture |
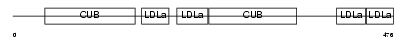
|
|||||
| Description | Low-density lipoprotein receptor-related protein 3 precursor (hLRp105). | |||||
|
LRP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.050229 (rank : 21) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75581 | Gene names | LRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 6 precursor. | |||||
|
LRP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.050597 (rank : 18) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88572 | Gene names | Lrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 6 precursor. | |||||
|
LRP8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.050312 (rank : 20) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14114, O14968, Q86V27, Q99876, Q9BR78 | Gene names | LRP8, APOER2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
LRP8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.050673 (rank : 17) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q924X6, Q8CAK9, Q8CDF5, Q921B6 | Gene names | Lrp8, Apoer2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
VLDLR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.051178 (rank : 14) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98155, Q5VVF6 | Gene names | VLDLR | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
VLDLR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.052675 (rank : 13) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98156, Q64022 | Gene names | Vldlr | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
SORC3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8VI51, Q9CTR4 | Gene names | Sorcs3 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
SORC3_HUMAN
|
||||||
| NC score | 0.994879 (rank : 2) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPU3, Q5VXF9, Q9NQJ2 | Gene names | SORCS3, KIAA1059 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
SORC1_HUMAN
|
||||||
| NC score | 0.993316 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WY21, Q59GG7, Q5JVT7, Q5JVT8, Q5VY14, Q86WQ1, Q86WQ2, Q9H1Y1, Q9H1Y2 | Gene names | SORCS1, SORCS | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS1 precursor (hSorCS). | |||||
|
SORC1_MOUSE
|
||||||
| NC score | 0.992814 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JLC4, Q8VI45, Q922I1, Q9QY21 | Gene names | Sorcs1, Sorcs | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS1 precursor (mSorCS). | |||||
|
SORC2_MOUSE
|
||||||
| NC score | 0.982754 (rank : 5) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EPR5 | Gene names | Sorcs2 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS2 precursor. | |||||
|
SORC2_HUMAN
|
||||||
| NC score | 0.978401 (rank : 6) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96PQ0, Q9P2L7 | Gene names | SORCS2, KIAA1329 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS2 precursor. | |||||
|
SORT_MOUSE
|
||||||
| NC score | 0.859378 (rank : 7) | θ value | 8.28633e-54 (rank : 10) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PHU5, Q3UHE2, Q8K043, Q9QXW6 | Gene names | Sort1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sortilin precursor (Neurotensin receptor 3) (NTR3) (mNTR3). | |||||
|
SORT_HUMAN
|
||||||
| NC score | 0.858637 (rank : 8) | θ value | 7.49467e-55 (rank : 9) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99523, Q8IZ49 | Gene names | SORT1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin precursor (Neurotensin receptor 3) (NTR3) (NT3) (Glycoprotein 95) (Gp95) (100 kDa NT receptor). | |||||
|
SORL_MOUSE
|
||||||
| NC score | 0.458969 (rank : 9) | θ value | 1.5555e-68 (rank : 7) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88307, O54711, O70581 | Gene names | Sorl1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (mSorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11) (Gp250). | |||||
|
SORL_HUMAN
|
||||||
| NC score | 0.455247 (rank : 10) | θ value | 7.71989e-68 (rank : 8) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92673, Q92856 | Gene names | SORL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (SorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11). | |||||
|
CD320_HUMAN
|
||||||
| NC score | 0.065857 (rank : 11) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NPF0, Q53HF7 | Gene names | CD320, 8D6A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD320 antigen precursor (8D6 antigen) (FDC-signaling molecule 8D6) (FDC-SM-8D6). | |||||
|
LRP2_HUMAN
|
||||||
| NC score | 0.054822 (rank : 12) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
VLDLR_MOUSE
|
||||||
| NC score | 0.052675 (rank : 13) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98156, Q64022 | Gene names | Vldlr | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
VLDLR_HUMAN
|
||||||
| NC score | 0.051178 (rank : 14) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98155, Q5VVF6 | Gene names | VLDLR | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
LRP3_HUMAN
|
||||||
| NC score | 0.051018 (rank : 15) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75074 | Gene names | LRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 3 precursor (hLRp105). | |||||
|
LRP12_MOUSE
|
||||||
| NC score | 0.050697 (rank : 16) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BUJ9, Q8BWM9 | Gene names | Lrp12 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor. | |||||
|
LRP8_MOUSE
|
||||||
| NC score | 0.050673 (rank : 17) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q924X6, Q8CAK9, Q8CDF5, Q921B6 | Gene names | Lrp8, Apoer2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
LRP6_MOUSE
|
||||||
| NC score | 0.050597 (rank : 18) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88572 | Gene names | Lrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 6 precursor. | |||||
|
LRP1_HUMAN
|
||||||
| NC score | 0.050398 (rank : 19) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
LRP8_HUMAN
|
||||||
| NC score | 0.050312 (rank : 20) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14114, O14968, Q86V27, Q99876, Q9BR78 | Gene names | LRP8, APOER2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
LRP6_HUMAN
|
||||||
| NC score | 0.050229 (rank : 21) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75581 | Gene names | LRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 6 precursor. | |||||
|
LDLR_MOUSE
|
||||||
| NC score | 0.050030 (rank : 22) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35951 | Gene names | Ldlr | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor precursor (LDL receptor). | |||||
|
PKD1_HUMAN
|
||||||
| NC score | 0.049884 (rank : 23) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
GCH1_HUMAN
|
||||||
| NC score | 0.048251 (rank : 24) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P30793, Q9Y4I8 | Gene names | GCH1, DYT5, GCH | |||
|
Domain Architecture |

|
|||||
| Description | GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I). | |||||
|
CUL7_HUMAN
|
||||||
| NC score | 0.033024 (rank : 25) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14999, Q5T654 | Gene names | CUL7, KIAA0076 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-7 (CUL-7). | |||||
|
T22D4_HUMAN
|
||||||
| NC score | 0.025120 (rank : 26) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.021811 (rank : 27) | θ value | 0.0431538 (rank : 11) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
PK1L1_HUMAN
|
||||||
| NC score | 0.020830 (rank : 28) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
PAPPA_HUMAN
|
||||||
| NC score | 0.020709 (rank : 29) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13219, Q08371, Q68G52, Q9UDK7 | Gene names | PAPPA | |||
|
Domain Architecture |

|
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.020638 (rank : 30) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
AOC3_HUMAN
|
||||||
| NC score | 0.019245 (rank : 31) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16853, Q45F94 | Gene names | AOC3, VAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane copper amine oxidase (EC 1.4.3.6) (Semicarbazide-sensitive amine oxidase) (SSAO) (Vascular adhesion protein 1) (VAP-1) (HPAO). | |||||
|
PHLD2_HUMAN
|
||||||
| NC score | 0.017541 (rank : 32) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15127 | Gene names | GPLD2, PIGPLD2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-glycan-specific phospholipase D 2 precursor (EC 3.1.4.50) (PI-G PLD) (Glycoprotein phospholipase D) (Glycosyl- phosphatidylinositol-specific phospholipase D). | |||||
|
GPNMB_HUMAN
|
||||||
| NC score | 0.016770 (rank : 33) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14956, Q6UVX1, Q8N1A1 | Gene names | GPNMB, HGFIN, NMB | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane glycoprotein NMB precursor (Transmembrane glycoprotein HGFIN). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.015968 (rank : 34) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
PME17_MOUSE
|
||||||
| NC score | 0.014740 (rank : 35) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60696 | Gene names | Silv, D10H12S53E, Pmel17, Si | |||
|
Domain Architecture |

|
|||||
| Description | Melanocyte protein Pmel 17 precursor (Silver locus protein). | |||||
|
GPR61_HUMAN
|
||||||
| NC score | 0.014551 (rank : 36) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZJ8, Q6NWS0, Q8TDV4, Q96PR4 | Gene names | GPR61, BALGR | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 61 (Biogenic amine receptor-like G-protein coupled receptor). | |||||
|
CT012_HUMAN
|
||||||
| NC score | 0.014409 (rank : 37) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NVP4, Q4F7X1, Q5QPD9, Q5QPE0, Q68DN8, Q6ZMX9, Q96NF0, Q9H1E0, Q9H442 | Gene names | C20orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C20orf12. | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.013763 (rank : 38) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
ZN541_HUMAN
|
||||||
| NC score | 0.013505 (rank : 39) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
PKHA4_HUMAN
|
||||||
| NC score | 0.012859 (rank : 40) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
GPR61_MOUSE
|
||||||
| NC score | 0.012605 (rank : 41) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 702 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C010 | Gene names | Gpr61 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 61. | |||||
|
ANR28_MOUSE
|
||||||
| NC score | 0.012130 (rank : 42) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q505D1 | Gene names | Ankrd28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR28_HUMAN
|
||||||
| NC score | 0.011843 (rank : 43) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15084, Q6ULS0, Q6ZT57 | Gene names | ANKRD28, KIAA0379 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
PRKDC_MOUSE
|
||||||
| NC score | 0.011322 (rank : 44) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97313, O88187, P97928, Q307W9, Q3V2W8, Q8C2A7, Q9Z341 | Gene names | Prkdc, Xrcc7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (P460). | |||||
|
CDYL1_HUMAN
|
||||||
| NC score | 0.011008 (rank : 45) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y232 | Gene names | CDYL, CDYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
PHLD1_HUMAN
|
||||||
| NC score | 0.010717 (rank : 46) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P80108, Q15128 | Gene names | GPLD1, PIGPLD1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-glycan-specific phospholipase D 1 precursor (EC 3.1.4.50) (PI-G PLD) (Glycoprotein phospholipase D) (Glycosyl- phosphatidylinositol-specific phospholipase D). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.009536 (rank : 47) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.008242 (rank : 48) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.007490 (rank : 49) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
FUCO2_HUMAN
|
||||||
| NC score | 0.006291 (rank : 50) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BTY2, Q7Z6Y2, Q8NBK4 | Gene names | FUCA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plasma alpha-L-fucosidase precursor (EC 3.2.1.51) (Alpha-L-fucosidase 2) (Alpha-L-fucoside fucohydrolase 2). | |||||
|
NLGN2_MOUSE
|
||||||
| NC score | 0.003350 (rank : 51) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q69ZK9, Q5F288 | Gene names | Nlgn2, Kiaa1366 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroligin-2 precursor. | |||||
|
WEE1_MOUSE
|
||||||
| NC score | 0.001506 (rank : 52) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 794 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47810, Q9EPL7 | Gene names | Wee1 | |||
|
Domain Architecture |

|
|||||
| Description | Wee1-like protein kinase (EC 2.7.10.2) (Wee1A kinase). | |||||
|
ZN219_HUMAN
|
||||||
| NC score | 0.000451 (rank : 53) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 876 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P2Y4, Q8IYC1, Q9BW28 | Gene names | ZNF219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 219. | |||||
|
AATK_HUMAN
|
||||||
| NC score | -0.000649 (rank : 54) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
MINK1_MOUSE
|
||||||
| NC score | -0.000872 (rank : 55) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1451 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JM52, Q921M6, Q9JM92 | Gene names | Mink1, Map4k6, Mink | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
MINK1_HUMAN
|
||||||
| NC score | -0.001101 (rank : 56) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N4C8, Q9P1X1, Q9P2R8 | Gene names | MINK1, MAP4K6, MINK | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
ZN206_HUMAN
|
||||||
| NC score | -0.001295 (rank : 57) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96SZ4 | Gene names | ZNF206, ZSCAN10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 206 (Zinc finger and SCAN domain-containing protein 10). | |||||