Please be patient as the page loads
|
PK1L1_HUMAN
|
||||||
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PK1L1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
PKDRE_HUMAN
|
||||||
| θ value | 1.28732e-30 (rank : 2) | NC score | 0.604179 (rank : 2) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PKD1_HUMAN
|
||||||
| θ value | 6.18819e-25 (rank : 3) | NC score | 0.356330 (rank : 9) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
PKDRE_MOUSE
|
||||||
| θ value | 3.07116e-24 (rank : 4) | NC score | 0.586976 (rank : 3) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PKD2_MOUSE
|
||||||
| θ value | 3.63628e-17 (rank : 5) | NC score | 0.504520 (rank : 4) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
PKD2_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 6) | NC score | 0.501822 (rank : 5) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
PK2L2_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 7) | NC score | 0.493587 (rank : 6) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L2_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 8) | NC score | 0.475296 (rank : 7) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L1_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 9) | NC score | 0.470979 (rank : 8) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
RA6I1_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 10) | NC score | 0.166795 (rank : 10) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6IQ26, Q96GN3, Q9H6U7, Q9UFV0, Q9UPR1 | Gene names | RAB6IP1, KIAA1091 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
RA6I1_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 11) | NC score | 0.162700 (rank : 11) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PAL8, Q62146, Q8C829, Q8VDF6, Q9QYZ2 | Gene names | Rab6ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
LOX5_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 12) | NC score | 0.108495 (rank : 12) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P09917 | Gene names | ALOX5, LOG5 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 5-lipoxygenase (EC 1.13.11.34) (5-lipoxygenase) (5-LO). | |||||
|
IF4G1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 13) | NC score | 0.074325 (rank : 23) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
ALO17_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 14) | NC score | 0.090321 (rank : 20) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 15) | NC score | 0.038400 (rank : 53) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 16) | NC score | 0.057986 (rank : 27) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
PME17_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 17) | NC score | 0.106574 (rank : 13) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60696 | Gene names | Silv, D10H12S53E, Pmel17, Si | |||
|
Domain Architecture |

|
|||||
| Description | Melanocyte protein Pmel 17 precursor (Silver locus protein). | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 18) | NC score | 0.064083 (rank : 26) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
CT2NL_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 19) | NC score | 0.044562 (rank : 43) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
IASPP_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 20) | NC score | 0.042275 (rank : 47) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 21) | NC score | 0.044998 (rank : 41) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.054008 (rank : 29) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
ARHGF_HUMAN
|
||||||
| θ value | 0.125558 (rank : 23) | NC score | 0.043391 (rank : 45) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O94989, Q8N449, Q9H8B4 | Gene names | ARHGEF15, KIAA0915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 15 (Vsm-RhoGEF). | |||||
|
DDFL1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 24) | NC score | 0.033738 (rank : 66) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TDY4, Q6P9F4, Q86UY1, Q9NXK2 | Gene names | DDEFL1, UPLC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1 (Protein up- regulated in liver cancer 1). | |||||
|
SYTL1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 25) | NC score | 0.032249 (rank : 71) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
NELFA_HUMAN
|
||||||
| θ value | 0.21417 (rank : 26) | NC score | 0.065330 (rank : 25) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H3P2, O95392 | Gene names | WHSC2, NELFA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 protein). | |||||
|
ABLM3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.028946 (rank : 81) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94929, Q658S1, Q68CI5, Q9BV32 | Gene names | ABLIM3, KIAA0843 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 3 (Actin-binding LIM protein family member 3) (abLIM-3). | |||||
|
LOX5_MOUSE
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.093196 (rank : 17) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P48999 | Gene names | Alox5 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 5-lipoxygenase (EC 1.13.11.34) (5-lipoxygenase) (5-LO). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.052602 (rank : 31) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.065866 (rank : 24) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
NU4M_MOUSE
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.051819 (rank : 33) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P03911 | Gene names | Mtnd4, mt-Nd4, Nd4 | |||
|
Domain Architecture |
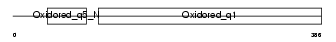
|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 4 (EC 1.6.5.3) (NADH dehydrogenase subunit 4). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.047764 (rank : 37) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
ABLM3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.027195 (rank : 85) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69ZX8, Q52KR1, Q6PAI7 | Gene names | Ablim3, Kiaa0843 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 3 (Actin-binding LIM protein family member 3) (abLIM-3). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.051110 (rank : 34) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
ICAM4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.047674 (rank : 38) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14773, Q14771, Q14772, Q16375 | Gene names | ICAM4, LW | |||
|
Domain Architecture |
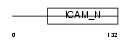
|
|||||
| Description | Intercellular adhesion molecule 4 precursor (ICAM-4) (Landsteiner- Wiener blood group glycoprotein) (LW blood group protein) (CD242 antigen). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.043457 (rank : 44) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PME17_HUMAN
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.086134 (rank : 22) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P40967, Q12763, Q14448, Q14817, Q16565 | Gene names | SILV, D12S53E, PMEL17 | |||
|
Domain Architecture |

|
|||||
| Description | Melanocyte protein Pmel 17 precursor (Melanocyte lineage-specific antigen GP100) (Melanoma-associated ME20 antigen) (ME20M/ME20S) (ME20- M/ME20-S) (95 kDa melanocyte-specific secreted glycoprotein). | |||||
|
RC3H1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.032892 (rank : 68) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
CENPT_MOUSE
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.050344 (rank : 35) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3TJM4, Q3TPY6, Q3UPD2, Q8BTH0, Q8BTP2, Q8R5E9 | Gene names | Cenpt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein T (CENP-T). | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.030529 (rank : 77) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
LIPE_HUMAN
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.040547 (rank : 50) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y5X9, Q6UW82 | Gene names | LIPG | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial lipase precursor (EC 3.1.1.3) (Endothelial cell-derived lipase) (EDL) (EL). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.042060 (rank : 49) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
TSC2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.034995 (rank : 62) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61037, P97723, P97724, P97725, P97727, Q61007, Q61008, Q9WUF6 | Gene names | Tsc2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 homolog protein). | |||||
|
ULK1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.008874 (rank : 133) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
LX12E_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.052976 (rank : 30) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P55249, Q91YW6 | Gene names | Alox12e, Alox12-ps2, Aloxe | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, epidermal-type (EC 1.13.11.31) (12-LOX). | |||||
|
LX12L_MOUSE
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.052160 (rank : 32) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P39654, Q4FJY9, Q5F2E3, Q6PHB2 | Gene names | Alox12l, Alox15 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, leukocyte-type (EC 1.13.11.31) (12-LOX). | |||||
|
RAI1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.044878 (rank : 42) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
ROBO3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.012370 (rank : 126) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96MS0 | Gene names | ROBO3 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Roundabout-like protein 3). | |||||
|
S39A4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.034676 (rank : 63) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q78IQ7, Q8CHL4 | Gene names | Slc39a4, Zip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc transporter ZIP4 precursor (Solute carrier family 39 member 4) (Activated in W/Wv mouse stomach 2) (mAWMS2). | |||||
|
AP3D1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.030240 (rank : 79) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.035642 (rank : 59) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.036728 (rank : 57) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.021773 (rank : 95) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
ARRD1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.030349 (rank : 78) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N5I2 | Gene names | ARRDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
|
ATX7_MOUSE
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.038925 (rank : 52) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.032674 (rank : 69) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CO3A1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.021214 (rank : 98) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
CSKI2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.019023 (rank : 105) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
F120A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.028652 (rank : 82) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZB2, O60649, Q14688, Q4VXF4, Q4VXF5, Q4VXG2, Q96I21, Q9NZB1 | Gene names | FAM120A, C9orf10, KIAA0183 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0318 protein FAM120A. | |||||
|
RUSC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.037331 (rank : 55) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N2Y8, O15080, Q6P1W7 | Gene names | RUSC2, KIAA0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
TARA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.030708 (rank : 76) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TMAP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.056685 (rank : 28) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CZH7, Q8C6F8 | Gene names | Tmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
CO3A1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.020750 (rank : 100) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
IF39_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.039923 (rank : 51) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
IF39_MOUSE
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.045336 (rank : 40) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
MK07_MOUSE
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.006098 (rank : 140) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1054 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WVS8 | Gene names | Mapk7, Erk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (BMK1 kinase). | |||||
|
SDK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.009233 (rank : 132) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q58EX2, Q86VY3, Q9NTD2, Q9NVB3, Q9P214 | Gene names | SDK2, KIAA1514 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-2 precursor. | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.042696 (rank : 46) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
ULK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.006941 (rank : 138) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75385 | Gene names | ULK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1). | |||||
|
ZEP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.012876 (rank : 125) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P31629, Q02646, Q5THT5, Q9NS05 | Gene names | HIVEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 (HIV- EP2) (MHC-binding protein 2) (MBP-2). | |||||
|
ZEP2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.013281 (rank : 124) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.008738 (rank : 134) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
DYN3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.016638 (rank : 116) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
IER2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.035759 (rank : 58) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BTL4, Q03827, Q2TAZ2 | Gene names | IER2, ETR101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immediate early response gene 2 protein (Protein ETR101). | |||||
|
K1267_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.026253 (rank : 87) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80TG1, Q3TT88, Q3U5D8, Q3V3N3, Q7TMU3, Q80XP7, Q8R3L6, Q9D9G0 | Gene names | Kiaa1267 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1267. | |||||
|
PO121_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.034998 (rank : 61) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
RTEL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.025743 (rank : 89) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NZ71, Q5JTV4, Q9BW37, Q9H402, Q9H4X6, Q9NX25, Q9NZ73, Q9UPR4, Q9Y4R6 | Gene names | RTEL1, C20orf41, KIAA1088, NHL | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of telomere elongation helicase 1 (EC 3.6.1.-) (Helicase- like protein NHL). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.028014 (rank : 83) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SLAF9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.020336 (rank : 102) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D780, Q8BTK0, Q8VE93 | Gene names | Slamf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SLAM family member 9 precursor. | |||||
|
TIF1B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.018049 (rank : 110) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13263, O00677, Q7Z632, Q93040, Q96IM1 | Gene names | TRIM28, KAP1, RNF96, TIF1B | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (Nuclear corepressor KAP-1) (KRAB- associated protein 1) (KAP-1) (KRAB-interacting protein 1) (KRIP-1) (RING finger protein 96). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.018336 (rank : 109) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
CCNK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.032550 (rank : 70) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CD2L7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.005214 (rank : 143) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
CDKL5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.002920 (rank : 145) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
FOXC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.010638 (rank : 129) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61572, O88409, Q61582 | Gene names | Foxc1, Fkh1, Fkhl7, Freac3, Mf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3) (Transcription factor FKH-1) (Mesoderm/mesenchyme forkhead 1) (MF-1). | |||||
|
GBX1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.006028 (rank : 141) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P82976, Q8CGW5 | Gene names | Gbx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein GBX-1 (Gastrulation and brain-specific homeobox protein 1). | |||||
|
K0226_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.027818 (rank : 84) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92622 | Gene names | KIAA0226 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0226. | |||||
|
L1CAM_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.007888 (rank : 137) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P32004, Q8TA87 | Gene names | L1CAM, CAML1, MIC5 | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule L1 precursor (N-CAM L1) (CD171 antigen). | |||||
|
LIPE_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.033864 (rank : 65) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WVG5, Q8VDU2 | Gene names | Lipg | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial lipase precursor (EC 3.1.1.3) (Endothelial cell-derived lipase) (EDL). | |||||
|
LPIN3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.016229 (rank : 118) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BQK8, Q5TDB9, Q9NPY8, Q9UJE5 | Gene names | LPIN3, LIPN3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-3 (Lipin 3-like). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.032175 (rank : 72) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.047308 (rank : 39) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
SORC3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.021699 (rank : 96) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPU3, Q5VXF9, Q9NQJ2 | Gene names | SORCS3, KIAA1059 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
WASF4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.037597 (rank : 54) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IV90 | Gene names | WASF4, SCAR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 4 (WASP-family protein member 4). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.049231 (rank : 36) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
COFA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.015778 (rank : 120) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35206, Q9EQD9 | Gene names | Col15a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV)]. | |||||
|
ELN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.023948 (rank : 92) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P54320 | Gene names | Eln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elastin precursor (Tropoelastin). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.031074 (rank : 73) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.033928 (rank : 64) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
RD23B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.024558 (rank : 91) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
RPGF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.016831 (rank : 114) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.017817 (rank : 112) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
SORC3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.020830 (rank : 99) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VI51, Q9CTR4 | Gene names | Sorcs3 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
AIM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.018657 (rank : 108) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y4K1, O00296, Q5VWJ2 | Gene names | AIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Absent in melanoma 1 protein. | |||||
|
AP3D1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.021999 (rank : 94) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
BASP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.030974 (rank : 74) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
EFNB3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.018866 (rank : 106) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15768, O00680, Q8TBH7, Q92875 | Gene names | EFNB3, EPLG8, LERK8 | |||
|
Domain Architecture |
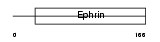
|
|||||
| Description | Ephrin-B3 precursor (EPH-related receptor tyrosine kinase ligand 8) (LERK-8) (EPH-related receptor transmembrane ligand ELK-L3). | |||||
|
EFNB3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.018675 (rank : 107) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35393 | Gene names | Efnb3 | |||
|
Domain Architecture |
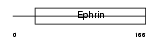
|
|||||
| Description | Ephrin-B3 precursor. | |||||
|
FRS2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.017931 (rank : 111) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C180 | Gene names | Frs2, Frs2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 2 (FGFR substrate 2) (Suc1-associated neurotrophic factor target 1) (SNT-1) (FGFR signaling adaptor SNT) (FRS2 alpha). | |||||
|
HXD11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.004393 (rank : 144) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31277, Q9NS02 | Gene names | HOXD11, HOX4F | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D11 (Hox-4F). | |||||
|
LNK_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.013984 (rank : 123) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UQQ2, O95184 | Gene names | LNK | |||
|
Domain Architecture |
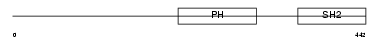
|
|||||
| Description | Lymphocyte-specific adapter protein Lnk (Signal transduction protein Lnk) (Lymphocyte adapter protein). | |||||
|
MEF2B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.017702 (rank : 113) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
MFRP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.005860 (rank : 142) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K480, Q8BPP4 | Gene names | Mfrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
MSH4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.012328 (rank : 127) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15457, Q8NEB3, Q9UNP8 | Gene names | MSH4 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 4. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.035470 (rank : 60) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PER1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.015903 (rank : 119) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.019642 (rank : 104) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
REXO1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.016822 (rank : 115) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TT28, Q3UMP1, Q69ZR0, Q6NSQ6, Q6PI95, Q9DA29 | Gene names | Rexo1, Kiaa1138, Tceb3bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
SYN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.025716 (rank : 90) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
TGM4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.008533 (rank : 135) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49221, Q16707, Q96QN4 | Gene names | TGM4 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase 4 (EC 2.3.2.13) (Transglutaminase-4) (TGase-4) (Prostate transglutaminase) (TGP) (TG(P)) (Prostate-specific transglutaminase) (Fibrinoligase). | |||||
|
TR19L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.015585 (rank : 121) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969Z4, Q86V34, Q96JU1, Q9BUX7 | Gene names | TNFRSF19L, RELT | |||
|
Domain Architecture |
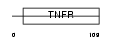
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 19L precursor (Receptor expressed in lymphoid tissues). | |||||
|
TRI56_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.011208 (rank : 128) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BRZ2, Q86VT6, Q8N2H8, Q8NAC0, Q9H031 | Gene names | TRIM56, RNF109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 56 (RING finger protein 109). | |||||
|
WASF2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.030878 (rank : 75) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
ZIMP7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.036880 (rank : 56) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NF64, O94790, Q659A8, Q6JKL5, Q8WTX8, Q96Q01, Q9BQH7 | Gene names | ZIMP7, KIAA1886 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
ANR47_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.008425 (rank : 136) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6NY19, Q6NZI1, Q6ZQR3, Q8IUV2 | Gene names | ANKRD47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 47. | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.020188 (rank : 103) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.021680 (rank : 97) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
E2F2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.010498 (rank : 130) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14209 | Gene names | E2F2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F2 (E2F-2). | |||||
|
FRS2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.016421 (rank : 117) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WU20, O43558, Q7LDQ6 | Gene names | FRS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 2 (FGFR substrate 2) (Suc1-associated neurotrophic factor target 1) (SNT-1) (FGFR signaling adaptor SNT). | |||||
|
GLI3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | -0.000500 (rank : 146) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61602 | Gene names | Gli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
KCNQ3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.006453 (rank : 139) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
LCP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.027054 (rank : 86) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.042129 (rank : 48) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.026147 (rank : 88) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
PO210_MOUSE
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.009502 (rank : 131) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QY81, Q3U031, Q69ZW2, Q7TQM1 | Gene names | Nup210, Kiaa0906 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore membrane glycoprotein 210 precursor (POM210) (Nuclear pore protein gp210). | |||||
|
PODXL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.020462 (rank : 101) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R0M4, Q9ESZ1 | Gene names | Podxl, Pclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.014544 (rank : 122) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
RUSC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.033150 (rank : 67) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.029833 (rank : 80) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.023808 (rank : 93) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
MCLN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.090379 (rank : 19) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9GZU1, Q7Z4F7, Q9H292, Q9H4B3, Q9H4B5 | Gene names | MCOLN1, ML4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-1 (Mucolipidin) (MG-2). | |||||
|
MCLN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.086983 (rank : 21) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99J21 | Gene names | Mcoln1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-1 (Mucolipidin). | |||||
|
MCLN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.093294 (rank : 16) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IZK6, Q8N9R3 | Gene names | MCOLN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
MCLN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.092766 (rank : 18) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K595, Q3UCG4, Q8K2T6, Q9CQD3 | Gene names | Mcoln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
MCLN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.097746 (rank : 14) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TDD5, Q5T4H5, Q5T4H6, Q9NV09 | Gene names | MCOLN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-3. | |||||
|
MCLN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.094555 (rank : 15) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R4F0, Q8BS73, Q8BSG1, Q8CDB2 | Gene names | Mcoln3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-3. | |||||
|
PK1L1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
PKDRE_HUMAN
|
||||||
| NC score | 0.604179 (rank : 2) | θ value | 1.28732e-30 (rank : 2) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PKDRE_MOUSE
|
||||||
| NC score | 0.586976 (rank : 3) | θ value | 3.07116e-24 (rank : 4) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PKD2_MOUSE
|
||||||
| NC score | 0.504520 (rank : 4) | θ value | 3.63628e-17 (rank : 5) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
PKD2_HUMAN
|
||||||
| NC score | 0.501822 (rank : 5) | θ value | 1.80466e-16 (rank : 6) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
PK2L2_MOUSE
|
||||||
| NC score | 0.493587 (rank : 6) | θ value | 3.40345e-15 (rank : 7) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L2_HUMAN
|
||||||
| NC score | 0.475296 (rank : 7) | θ value | 4.59992e-12 (rank : 8) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L1_HUMAN
|
||||||
| NC score | 0.470979 (rank : 8) | θ value | 1.63604e-09 (rank : 9) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PKD1_HUMAN
|
||||||
| NC score | 0.356330 (rank : 9) | θ value | 6.18819e-25 (rank : 3) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
RA6I1_HUMAN
|
||||||
| NC score | 0.166795 (rank : 10) | θ value | 5.44631e-05 (rank : 10) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6IQ26, Q96GN3, Q9H6U7, Q9UFV0, Q9UPR1 | Gene names | RAB6IP1, KIAA1091 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
RA6I1_MOUSE
|
||||||
| NC score | 0.162700 (rank : 11) | θ value | 9.29e-05 (rank : 11) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PAL8, Q62146, Q8C829, Q8VDF6, Q9QYZ2 | Gene names | Rab6ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
LOX5_HUMAN
|
||||||
| NC score | 0.108495 (rank : 12) | θ value | 0.00390308 (rank : 12) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P09917 | Gene names | ALOX5, LOG5 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 5-lipoxygenase (EC 1.13.11.34) (5-lipoxygenase) (5-LO). | |||||
|
PME17_MOUSE
|
||||||
| NC score | 0.106574 (rank : 13) | θ value | 0.0252991 (rank : 17) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60696 | Gene names | Silv, D10H12S53E, Pmel17, Si | |||
|
Domain Architecture |

|
|||||
| Description | Melanocyte protein Pmel 17 precursor (Silver locus protein). | |||||
|
MCLN3_HUMAN
|
||||||
| NC score | 0.097746 (rank : 14) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TDD5, Q5T4H5, Q5T4H6, Q9NV09 | Gene names | MCOLN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-3. | |||||
|
MCLN3_MOUSE
|
||||||
| NC score | 0.094555 (rank : 15) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R4F0, Q8BS73, Q8BSG1, Q8CDB2 | Gene names | Mcoln3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-3. | |||||
|
MCLN2_HUMAN
|
||||||
| NC score | 0.093294 (rank : 16) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IZK6, Q8N9R3 | Gene names | MCOLN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
LOX5_MOUSE
|
||||||
| NC score | 0.093196 (rank : 17) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P48999 | Gene names | Alox5 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 5-lipoxygenase (EC 1.13.11.34) (5-lipoxygenase) (5-LO). | |||||
|
MCLN2_MOUSE
|
||||||
| NC score | 0.092766 (rank : 18) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K595, Q3UCG4, Q8K2T6, Q9CQD3 | Gene names | Mcoln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
MCLN1_HUMAN
|
||||||
| NC score | 0.090379 (rank : 19) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9GZU1, Q7Z4F7, Q9H292, Q9H4B3, Q9H4B5 | Gene names | MCOLN1, ML4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-1 (Mucolipidin) (MG-2). | |||||
|
ALO17_HUMAN
|
||||||
| NC score | 0.090321 (rank : 20) | θ value | 0.0252991 (rank : 14) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
MCLN1_MOUSE
|
||||||
| NC score | 0.086983 (rank : 21) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99J21 | Gene names | Mcoln1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-1 (Mucolipidin). | |||||
|
PME17_HUMAN
|
||||||
| NC score | 0.086134 (rank : 22) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P40967, Q12763, Q14448, Q14817, Q16565 | Gene names | SILV, D12S53E, PMEL17 | |||
|
Domain Architecture |

|
|||||
| Description | Melanocyte protein Pmel 17 precursor (Melanocyte lineage-specific antigen GP100) (Melanoma-associated ME20 antigen) (ME20M/ME20S) (ME20- M/ME20-S) (95 kDa melanocyte-specific secreted glycoprotein). | |||||
|
IF4G1_HUMAN
|
||||||
| NC score | 0.074325 (rank : 23) | θ value | 0.0113563 (rank : 13) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.065866 (rank : 24) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
NELFA_HUMAN
|
||||||
| NC score | 0.065330 (rank : 25) | θ value | 0.21417 (rank : 26) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H3P2, O95392 | Gene names | WHSC2, NELFA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 protein). | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.064083 (rank : 26) | θ value | 0.0330416 (rank : 18) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.057986 (rank : 27) | θ value | 0.0252991 (rank : 16) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
TMAP1_MOUSE
|
||||||
| NC score | 0.056685 (rank : 28) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CZH7, Q8C6F8 | Gene names | Tmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.054008 (rank : 29) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
LX12E_MOUSE
|
||||||
| NC score | 0.052976 (rank : 30) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P55249, Q91YW6 | Gene names | Alox12e, Alox12-ps2, Aloxe | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, epidermal-type (EC 1.13.11.31) (12-LOX). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.052602 (rank : 31) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
LX12L_MOUSE
|
||||||
| NC score | 0.052160 (rank : 32) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P39654, Q4FJY9, Q5F2E3, Q6PHB2 | Gene names | Alox12l, Alox15 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, leukocyte-type (EC 1.13.11.31) (12-LOX). | |||||
|
NU4M_MOUSE
|
||||||
| NC score | 0.051819 (rank : 33) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P03911 | Gene names | Mtnd4, mt-Nd4, Nd4 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 4 (EC 1.6.5.3) (NADH dehydrogenase subunit 4). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.051110 (rank : 34) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CENPT_MOUSE
|
||||||
| NC score | 0.050344 (rank : 35) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3TJM4, Q3TPY6, Q3UPD2, Q8BTH0, Q8BTP2, Q8R5E9 | Gene names | Cenpt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein T (CENP-T). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.049231 (rank : 36) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.047764 (rank : 37) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
ICAM4_HUMAN
|
||||||
| NC score | 0.047674 (rank : 38) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14773, Q14771, Q14772, Q16375 | Gene names | ICAM4, LW | |||
|
Domain Architecture |

|
|||||
| Description | Intercellular adhesion molecule 4 precursor (ICAM-4) (Landsteiner- Wiener blood group glycoprotein) (LW blood group protein) (CD242 antigen). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.047308 (rank : 39) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
IF39_MOUSE
|
||||||
| NC score | 0.045336 (rank : 40) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.044998 (rank : 41) | θ value | 0.0961366 (rank : 21) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.044878 (rank : 42) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
CT2NL_MOUSE
|
||||||
| NC score | 0.044562 (rank : 43) | θ value | 0.0563607 (rank : 19) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.043457 (rank : 44) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
ARHGF_HUMAN
|
||||||
| NC score | 0.043391 (rank : 45) | θ value | 0.125558 (rank : 23) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O94989, Q8N449, Q9H8B4 | Gene names | ARHGEF15, KIAA0915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 15 (Vsm-RhoGEF). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.042696 (rank : 46) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
IASPP_HUMAN
|
||||||
| NC score | 0.042275 (rank : 47) | θ value | 0.0563607 (rank : 20) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.042129 (rank : 48) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.042060 (rank : 49) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
LIPE_HUMAN
|
||||||
| NC score | 0.040547 (rank : 50) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y5X9, Q6UW82 | Gene names | LIPG | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial lipase precursor (EC 3.1.1.3) (Endothelial cell-derived lipase) (EDL) (EL). | |||||
|
IF39_HUMAN
|
||||||
| NC score | 0.039923 (rank : 51) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
ATX7_MOUSE
|
||||||
| NC score | 0.038925 (rank : 52) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.038400 (rank : 53) | θ value | 0.0252991 (rank : 15) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
WASF4_HUMAN
|
||||||
| NC score | 0.037597 (rank : 54) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IV90 | Gene names | WASF4, SCAR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 4 (WASP-family protein member 4). | |||||
|
RUSC2_HUMAN
|
||||||
| NC score | 0.037331 (rank : 55) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N2Y8, O15080, Q6P1W7 | Gene names | RUSC2, KIAA0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
ZIMP7_HUMAN
|
||||||
| NC score | 0.036880 (rank : 56) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NF64, O94790, Q659A8, Q6JKL5, Q8WTX8, Q96Q01, Q9BQH7 | Gene names | ZIMP7, KIAA1886 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.036728 (rank : 57) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
IER2_HUMAN
|
||||||
| NC score | 0.035759 (rank : 58) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BTL4, Q03827, Q2TAZ2 | Gene names | IER2, ETR101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immediate early response gene 2 protein (Protein ETR101). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.035642 (rank : 59) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.035470 (rank : 60) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.034998 (rank : 61) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
TSC2_MOUSE
|
||||||
| NC score | 0.034995 (rank : 62) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61037, P97723, P97724, P97725, P97727, Q61007, Q61008, Q9WUF6 | Gene names | Tsc2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 homolog protein). | |||||
|
S39A4_MOUSE
|
||||||
| NC score | 0.034676 (rank : 63) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q78IQ7, Q8CHL4 | Gene names | Slc39a4, Zip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc transporter ZIP4 precursor (Solute carrier family 39 member 4) (Activated in W/Wv mouse stomach 2) (mAWMS2). | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.033928 (rank : 64) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
LIPE_MOUSE
|
||||||
| NC score | 0.033864 (rank : 65) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WVG5, Q8VDU2 | Gene names | Lipg | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial lipase precursor (EC 3.1.1.3) (Endothelial cell-derived lipase) (EDL). | |||||
|
DDFL1_HUMAN
|
||||||
| NC score | 0.033738 (rank : 66) | θ value | 0.125558 (rank : 24) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TDY4, Q6P9F4, Q86UY1, Q9NXK2 | Gene names | DDEFL1, UPLC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1 (Protein up- regulated in liver cancer 1). | |||||
|
RUSC2_MOUSE
|
||||||
| NC score | 0.033150 (rank : 67) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
RC3H1_HUMAN
|
||||||
| NC score | 0.032892 (rank : 68) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.032674 (rank : 69) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.032550 (rank : 70) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
SYTL1_MOUSE
|
||||||
| NC score | 0.032249 (rank : 71) | θ value | 0.125558 (rank : 25) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.032175 (rank : 72) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.031074 (rank : 73) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.030974 (rank : 74) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
WASF2_HUMAN
|
||||||
| NC score | 0.030878 (rank : 75) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.030708 (rank : 76) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.030529 (rank : 77) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
ARRD1_HUMAN
|
||||||
| NC score | 0.030349 (rank : 78) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N5I2 | Gene names | ARRDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
|
AP3D1_MOUSE
|
||||||
| NC score | 0.030240 (rank : 79) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.029833 (rank : 80) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
ABLM3_HUMAN
|
||||||
| NC score | 0.028946 (rank : 81) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94929, Q658S1, Q68CI5, Q9BV32 | Gene names | ABLIM3, KIAA0843 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 3 (Actin-binding LIM protein family member 3) (abLIM-3). | |||||
|
F120A_HUMAN
|
||||||
| NC score | 0.028652 (rank : 82) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZB2, O60649, Q14688, Q4VXF4, Q4VXF5, Q4VXG2, Q96I21, Q9NZB1 | Gene names | FAM120A, C9orf10, KIAA0183 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0318 protein FAM120A. | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.028014 (rank : 83) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
K0226_HUMAN
|
||||||
| NC score | 0.027818 (rank : 84) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92622 | Gene names | KIAA0226 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0226. | |||||
|
ABLM3_MOUSE
|
||||||
| NC score | 0.027195 (rank : 85) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69ZX8, Q52KR1, Q6PAI7 | Gene names | Ablim3, Kiaa0843 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 3 (Actin-binding LIM protein family member 3) (abLIM-3). | |||||
|
LCP2_HUMAN
|
||||||
| NC score | 0.027054 (rank : 86) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
K1267_MOUSE
|
||||||
| NC score | 0.026253 (rank : 87) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80TG1, Q3TT88, Q3U5D8, Q3V3N3, Q7TMU3, Q80XP7, Q8R3L6, Q9D9G0 | Gene names | Kiaa1267 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1267. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.026147 (rank : 88) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
RTEL1_HUMAN
|
||||||
| NC score | 0.025743 (rank : 89) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NZ71, Q5JTV4, Q9BW37, Q9H402, Q9H4X6, Q9NX25, Q9NZ73, Q9UPR4, Q9Y4R6 | Gene names | RTEL1, C20orf41, KIAA1088, NHL | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of telomere elongation helicase 1 (EC 3.6.1.-) (Helicase- like protein NHL). | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.025716 (rank : 90) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
RD23B_MOUSE
|
||||||
| NC score | 0.024558 (rank : 91) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
ELN_MOUSE
|
||||||
| NC score | 0.023948 (rank : 92) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P54320 | Gene names | Eln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elastin precursor (Tropoelastin). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.023808 (rank : 93) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.021999 (rank : 94) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.021773 (rank : 95) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
SORC3_HUMAN
|
||||||
| NC score | 0.021699 (rank : 96) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPU3, Q5VXF9, Q9NQJ2 | Gene names | SORCS3, KIAA1059 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.021680 (rank : 97) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
CO3A1_MOUSE
|
||||||
| NC score | 0.021214 (rank : 98) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
SORC3_MOUSE
|
||||||
| NC score | 0.020830 (rank : 99) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VI51, Q9CTR4 | Gene names | Sorcs3 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
CO3A1_HUMAN
|
||||||
| NC score | 0.020750 (rank : 100) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
PODXL_MOUSE
|
||||||
| NC score | 0.020462 (rank : 101) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R0M4, Q9ESZ1 | Gene names | Podxl, Pclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
SLAF9_MOUSE
|
||||||
| NC score | 0.020336 (rank : 102) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D780, Q8BTK0, Q8VE93 | Gene names | Slamf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SLAM family member 9 precursor. | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.020188 (rank : 103) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.019642 (rank : 104) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
CSKI2_HUMAN
|
||||||
| NC score | 0.019023 (rank : 105) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
EFNB3_HUMAN
|
||||||
| NC score | 0.018866 (rank : 106) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15768, O00680, Q8TBH7, Q92875 | Gene names | EFNB3, EPLG8, LERK8 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin-B3 precursor (EPH-related receptor tyrosine kinase ligand 8) (LERK-8) (EPH-related receptor transmembrane ligand ELK-L3). | |||||
|
EFNB3_MOUSE
|
||||||
| NC score | 0.018675 (rank : 107) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35393 | Gene names | Efnb3 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin-B3 precursor. | |||||
|
AIM1_HUMAN
|
||||||
| NC score | 0.018657 (rank : 108) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y4K1, O00296, Q5VWJ2 | Gene names | AIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Absent in melanoma 1 protein. | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.018336 (rank : 109) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
TIF1B_HUMAN
|
||||||
| NC score | 0.018049 (rank : 110) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13263, O00677, Q7Z632, Q93040, Q96IM1 | Gene names | TRIM28, KAP1, RNF96, TIF1B | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (Nuclear corepressor KAP-1) (KRAB- associated protein 1) (KAP-1) (KRAB-interacting protein 1) (KRIP-1) (RING finger protein 96). | |||||
|
FRS2_MOUSE
|
||||||
| NC score | 0.017931 (rank : 111) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C180 | Gene names | Frs2, Frs2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 2 (FGFR substrate 2) (Suc1-associated neurotrophic factor target 1) (SNT-1) (FGFR signaling adaptor SNT) (FRS2 alpha). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.017817 (rank : 112) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
MEF2B_MOUSE
|
||||||
| NC score | 0.017702 (rank : 113) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
RPGF1_HUMAN
|
||||||
| NC score | 0.016831 (rank : 114) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
REXO1_MOUSE
|
||||||
| NC score | 0.016822 (rank : 115) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TT28, Q3UMP1, Q69ZR0, Q6NSQ6, Q6PI95, Q9DA29 | Gene names | Rexo1, Kiaa1138, Tceb3bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
DYN3_HUMAN
|
||||||
| NC score | 0.016638 (rank : 116) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
FRS2_HUMAN
|
||||||
| NC score | 0.016421 (rank : 117) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WU20, O43558, Q7LDQ6 | Gene names | FRS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 2 (FGFR substrate 2) (Suc1-associated neurotrophic factor target 1) (SNT-1) (FGFR signaling adaptor SNT). | |||||
|
LPIN3_HUMAN
|
||||||
| NC score | 0.016229 (rank : 118) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BQK8, Q5TDB9, Q9NPY8, Q9UJE5 | Gene names | LPIN3, LIPN3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-3 (Lipin 3-like). | |||||
|
PER1_MOUSE
|
||||||
| NC score | 0.015903 (rank : 119) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
COFA1_MOUSE
|
||||||
| NC score | 0.015778 (rank : 120) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35206, Q9EQD9 | Gene names | Col15a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV)]. | |||||
|
TR19L_HUMAN
|
||||||
| NC score | 0.015585 (rank : 121) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969Z4, Q86V34, Q96JU1, Q9BUX7 | Gene names | TNFRSF19L, RELT | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 19L precursor (Receptor expressed in lymphoid tissues). | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.014544 (rank : 122) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
LNK_HUMAN
|
||||||
| NC score | 0.013984 (rank : 123) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UQQ2, O95184 | Gene names | LNK | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte-specific adapter protein Lnk (Signal transduction protein Lnk) (Lymphocyte adapter protein). | |||||
|
ZEP2_MOUSE
|
||||||
| NC score | 0.013281 (rank : 124) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
ZEP2_HUMAN
|
||||||
| NC score | 0.012876 (rank : 125) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P31629, Q02646, Q5THT5, Q9NS05 | Gene names | HIVEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 (HIV- EP2) (MHC-binding protein 2) (MBP-2). | |||||
|
ROBO3_HUMAN
|
||||||
| NC score | 0.012370 (rank : 126) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96MS0 | Gene names | ROBO3 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Roundabout-like protein 3). | |||||
|
MSH4_HUMAN
|
||||||
| NC score | 0.012328 (rank : 127) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15457, Q8NEB3, Q9UNP8 | Gene names | MSH4 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 4. | |||||
|
TRI56_HUMAN
|
||||||
| NC score | 0.011208 (rank : 128) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BRZ2, Q86VT6, Q8N2H8, Q8NAC0, Q9H031 | Gene names | TRIM56, RNF109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 56 (RING finger protein 109). | |||||
|
FOXC1_MOUSE
|
||||||
| NC score | 0.010638 (rank : 129) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61572, O88409, Q61582 | Gene names | Foxc1, Fkh1, Fkhl7, Freac3, Mf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3) (Transcription factor FKH-1) (Mesoderm/mesenchyme forkhead 1) (MF-1). | |||||
|
E2F2_HUMAN
|
||||||
| NC score | 0.010498 (rank : 130) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14209 | Gene names | E2F2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F2 (E2F-2). | |||||
|
PO210_MOUSE
|
||||||
| NC score | 0.009502 (rank : 131) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QY81, Q3U031, Q69ZW2, Q7TQM1 | Gene names | Nup210, Kiaa0906 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore membrane glycoprotein 210 precursor (POM210) (Nuclear pore protein gp210). | |||||
|
SDK2_HUMAN
|
||||||
| NC score | 0.009233 (rank : 132) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q58EX2, Q86VY3, Q9NTD2, Q9NVB3, Q9P214 | Gene names | SDK2, KIAA1514 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-2 precursor. | |||||
|
ULK1_MOUSE
|
||||||
| NC score | 0.008874 (rank : 133) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.008738 (rank : 134) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
TGM4_HUMAN
|
||||||
| NC score | 0.008533 (rank : 135) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49221, Q16707, Q96QN4 | Gene names | TGM4 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase 4 (EC 2.3.2.13) (Transglutaminase-4) (TGase-4) (Prostate transglutaminase) (TGP) (TG(P)) (Prostate-specific transglutaminase) (Fibrinoligase). | |||||
|
ANR47_HUMAN
|
||||||
| NC score | 0.008425 (rank : 136) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6NY19, Q6NZI1, Q6ZQR3, Q8IUV2 | Gene names | ANKRD47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 47. | |||||
|
L1CAM_HUMAN
|
||||||
| NC score | 0.007888 (rank : 137) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P32004, Q8TA87 | Gene names | L1CAM, CAML1, MIC5 | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule L1 precursor (N-CAM L1) (CD171 antigen). | |||||
|
ULK1_HUMAN
|
||||||
| NC score | 0.006941 (rank : 138) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75385 | Gene names | ULK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1). | |||||
|
KCNQ3_MOUSE
|
||||||
| NC score | 0.006453 (rank : 139) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
MK07_MOUSE
|
||||||
| NC score | 0.006098 (rank : 140) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1054 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WVS8 | Gene names | Mapk7, Erk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (BMK1 kinase). | |||||
|
GBX1_MOUSE
|
||||||
| NC score | 0.006028 (rank : 141) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P82976, Q8CGW5 | Gene names | Gbx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein GBX-1 (Gastrulation and brain-specific homeobox protein 1). | |||||
|
MFRP_MOUSE
|
||||||
| NC score | 0.005860 (rank : 142) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K480, Q8BPP4 | Gene names | Mfrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
CD2L7_HUMAN
|
||||||
| NC score | 0.005214 (rank : 143) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
HXD11_HUMAN
|
||||||
| NC score | 0.004393 (rank : 144) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31277, Q9NS02 | Gene names | HOXD11, HOX4F | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D11 (Hox-4F). | |||||
|
CDKL5_HUMAN
|
||||||
| NC score | 0.002920 (rank : 145) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
GLI3_MOUSE
|
||||||
| NC score | -0.000500 (rank : 146) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61602 | Gene names | Gli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||