Please be patient as the page loads
|
NU4M_MOUSE
|
||||||
| SwissProt Accessions | P03911 | Gene names | Mtnd4, mt-Nd4, Nd4 | |||
|
Domain Architecture |
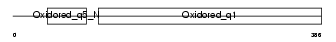
|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 4 (EC 1.6.5.3) (NADH dehydrogenase subunit 4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NU4M_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P03911 | Gene names | Mtnd4, mt-Nd4, Nd4 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 4 (EC 1.6.5.3) (NADH dehydrogenase subunit 4). | |||||
|
NU4M_HUMAN
|
||||||
| θ value | 1.05009e-157 (rank : 2) | NC score | 0.991318 (rank : 2) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P03905, Q6RL39, Q6RQN9, Q8HNR8 | Gene names | MT-ND4, MTND4, NADH4, ND4 | |||
|
Domain Architecture |
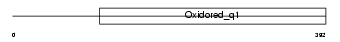
|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 4 (EC 1.6.5.3) (NADH dehydrogenase subunit 4). | |||||
|
NU5M_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 3) | NC score | 0.581520 (rank : 3) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03921 | Gene names | Mtnd5, mt-Nd5, Nd5 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 5 (EC 1.6.5.3) (NADH dehydrogenase subunit 5). | |||||
|
NU5M_HUMAN
|
||||||
| θ value | 2.98157e-11 (rank : 4) | NC score | 0.535518 (rank : 4) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03915, Q34773, Q8WCY3 | Gene names | MT-ND5, MTND5, NADH5, ND5 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 5 (EC 1.6.5.3) (NADH dehydrogenase subunit 5). | |||||
|
NU2M_MOUSE
|
||||||
| θ value | 8.67504e-11 (rank : 5) | NC score | 0.534007 (rank : 5) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03893 | Gene names | Mtnd2, mt-Nd2, Nd2 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 2 (EC 1.6.5.3) (NADH dehydrogenase subunit 2). | |||||
|
NU2M_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 6) | NC score | 0.469977 (rank : 6) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03891, Q34769, Q9TGI0, Q9TGI1, Q9TGI2, Q9TGI3, Q9TGI4 | Gene names | MT-ND2, MTND2, NADH2, ND2 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 2 (EC 1.6.5.3) (NADH dehydrogenase subunit 2). | |||||
|
PK1L1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 7) | NC score | 0.051819 (rank : 7) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
O1102_MOUSE
|
||||||
| θ value | 0.62314 (rank : 8) | NC score | 0.005472 (rank : 13) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 630 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VES2 | Gene names | Olfr1102, Mor179-4 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 1102 (Olfactory receptor 179-4). | |||||
|
OR1E1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 9) | NC score | 0.004356 (rank : 15) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P30953, O43884, P47882, P47885, Q9UBJ1, Q9UM60 | Gene names | OR1E1, OR1E5, OR1E6 | |||
|
Domain Architecture |
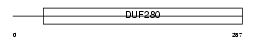
|
|||||
| Description | Olfactory receptor 1E1 (Olfactory receptor-like protein HGMP07I) (Olfactory receptor 17-2/17-32) (OR17-2) (OR17-32) (Olfactory receptor 13-66) (OR13-66) (Olfactory receptor 5-85) (OR5-85). | |||||
|
SC6A2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 10) | NC score | 0.015091 (rank : 10) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O55192 | Gene names | Slc6a2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium-dependent noradrenaline transporter (Norepinephrine transporter) (NET). | |||||
|
SC6A2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 11) | NC score | 0.013508 (rank : 11) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23975 | Gene names | SLC6A2, NAT1, NET1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium-dependent noradrenaline transporter (Norepinephrine transporter) (NET). | |||||
|
ASAH3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 12) | NC score | 0.017641 (rank : 8) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R4X1 | Gene names | Asah3, Acer1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkaline ceramidase 1 (EC 3.5.1.23) (Alkaline CDase-1) (AlkCDase 1) (maCER1) (Acylsphingosine deacylase 3) (N-acylsphingosine amidohydrolase 3). | |||||
|
O2T12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 13) | NC score | 0.004715 (rank : 14) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 656 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NG77 | Gene names | OR2T12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 2T12 (Olfactory receptor OR1-57). | |||||
|
PSN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 14) | NC score | 0.015386 (rank : 9) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49810, Q96P32 | Gene names | PSEN2, AD4, PS2, PSNL2, STM2 | |||
|
Domain Architecture |
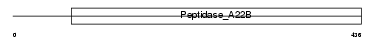
|
|||||
| Description | Presenilin-2 (EC 3.4.23.-) (PS-2) (STM-2) (E5-1) (AD3LP) (AD5) [Contains: Presenilin-2 NTF subunit; Presenilin-2 CTF subunit]. | |||||
|
TR125_MOUSE
|
||||||
| θ value | 8.99809 (rank : 15) | NC score | 0.010577 (rank : 12) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7M710 | Gene names | Tas2r125, T2r59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 2 member 125 (T2R125) (mT2R59). | |||||
|
NU4M_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P03911 | Gene names | Mtnd4, mt-Nd4, Nd4 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 4 (EC 1.6.5.3) (NADH dehydrogenase subunit 4). | |||||
|
NU4M_HUMAN
|
||||||
| NC score | 0.991318 (rank : 2) | θ value | 1.05009e-157 (rank : 2) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P03905, Q6RL39, Q6RQN9, Q8HNR8 | Gene names | MT-ND4, MTND4, NADH4, ND4 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 4 (EC 1.6.5.3) (NADH dehydrogenase subunit 4). | |||||
|
NU5M_MOUSE
|
||||||
| NC score | 0.581520 (rank : 3) | θ value | 3.40345e-15 (rank : 3) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03921 | Gene names | Mtnd5, mt-Nd5, Nd5 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 5 (EC 1.6.5.3) (NADH dehydrogenase subunit 5). | |||||
|
NU5M_HUMAN
|
||||||
| NC score | 0.535518 (rank : 4) | θ value | 2.98157e-11 (rank : 4) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03915, Q34773, Q8WCY3 | Gene names | MT-ND5, MTND5, NADH5, ND5 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 5 (EC 1.6.5.3) (NADH dehydrogenase subunit 5). | |||||
|
NU2M_MOUSE
|
||||||
| NC score | 0.534007 (rank : 5) | θ value | 8.67504e-11 (rank : 5) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03893 | Gene names | Mtnd2, mt-Nd2, Nd2 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 2 (EC 1.6.5.3) (NADH dehydrogenase subunit 2). | |||||
|
NU2M_HUMAN
|
||||||
| NC score | 0.469977 (rank : 6) | θ value | 6.43352e-06 (rank : 6) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03891, Q34769, Q9TGI0, Q9TGI1, Q9TGI2, Q9TGI3, Q9TGI4 | Gene names | MT-ND2, MTND2, NADH2, ND2 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 2 (EC 1.6.5.3) (NADH dehydrogenase subunit 2). | |||||
|
PK1L1_HUMAN
|
||||||
| NC score | 0.051819 (rank : 7) | θ value | 0.47712 (rank : 7) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
ASAH3_MOUSE
|
||||||
| NC score | 0.017641 (rank : 8) | θ value | 6.88961 (rank : 12) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R4X1 | Gene names | Asah3, Acer1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkaline ceramidase 1 (EC 3.5.1.23) (Alkaline CDase-1) (AlkCDase 1) (maCER1) (Acylsphingosine deacylase 3) (N-acylsphingosine amidohydrolase 3). | |||||
|
PSN2_HUMAN
|
||||||
| NC score | 0.015386 (rank : 9) | θ value | 6.88961 (rank : 14) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49810, Q96P32 | Gene names | PSEN2, AD4, PS2, PSNL2, STM2 | |||
|
Domain Architecture |

|
|||||
| Description | Presenilin-2 (EC 3.4.23.-) (PS-2) (STM-2) (E5-1) (AD3LP) (AD5) [Contains: Presenilin-2 NTF subunit; Presenilin-2 CTF subunit]. | |||||
|
SC6A2_MOUSE
|
||||||
| NC score | 0.015091 (rank : 10) | θ value | 1.81305 (rank : 10) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O55192 | Gene names | Slc6a2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium-dependent noradrenaline transporter (Norepinephrine transporter) (NET). | |||||
|
SC6A2_HUMAN
|
||||||
| NC score | 0.013508 (rank : 11) | θ value | 4.03905 (rank : 11) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23975 | Gene names | SLC6A2, NAT1, NET1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium-dependent noradrenaline transporter (Norepinephrine transporter) (NET). | |||||
|
TR125_MOUSE
|
||||||
| NC score | 0.010577 (rank : 12) | θ value | 8.99809 (rank : 15) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7M710 | Gene names | Tas2r125, T2r59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 2 member 125 (T2R125) (mT2R59). | |||||
|
O1102_MOUSE
|
||||||
| NC score | 0.005472 (rank : 13) | θ value | 0.62314 (rank : 8) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 630 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VES2 | Gene names | Olfr1102, Mor179-4 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 1102 (Olfactory receptor 179-4). | |||||
|
O2T12_HUMAN
|
||||||
| NC score | 0.004715 (rank : 14) | θ value | 6.88961 (rank : 13) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 656 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NG77 | Gene names | OR2T12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 2T12 (Olfactory receptor OR1-57). | |||||
|
OR1E1_HUMAN
|
||||||
| NC score | 0.004356 (rank : 15) | θ value | 1.81305 (rank : 9) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P30953, O43884, P47882, P47885, Q9UBJ1, Q9UM60 | Gene names | OR1E1, OR1E5, OR1E6 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 1E1 (Olfactory receptor-like protein HGMP07I) (Olfactory receptor 17-2/17-32) (OR17-2) (OR17-32) (Olfactory receptor 13-66) (OR13-66) (Olfactory receptor 5-85) (OR5-85). | |||||