Please be patient as the page loads
|
NU5M_HUMAN
|
||||||
| SwissProt Accessions | P03915, Q34773, Q8WCY3 | Gene names | MT-ND5, MTND5, NADH5, ND5 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 5 (EC 1.6.5.3) (NADH dehydrogenase subunit 5). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NU5M_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P03915, Q34773, Q8WCY3 | Gene names | MT-ND5, MTND5, NADH5, ND5 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 5 (EC 1.6.5.3) (NADH dehydrogenase subunit 5). | |||||
|
NU5M_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.990702 (rank : 2) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03921 | Gene names | Mtnd5, mt-Nd5, Nd5 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 5 (EC 1.6.5.3) (NADH dehydrogenase subunit 5). | |||||
|
NU2M_MOUSE
|
||||||
| θ value | 2.28291e-11 (rank : 3) | NC score | 0.551802 (rank : 3) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03893 | Gene names | Mtnd2, mt-Nd2, Nd2 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 2 (EC 1.6.5.3) (NADH dehydrogenase subunit 2). | |||||
|
NU4M_MOUSE
|
||||||
| θ value | 2.98157e-11 (rank : 4) | NC score | 0.535518 (rank : 4) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03911 | Gene names | Mtnd4, mt-Nd4, Nd4 | |||
|
Domain Architecture |
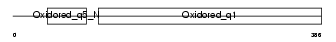
|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 4 (EC 1.6.5.3) (NADH dehydrogenase subunit 4). | |||||
|
NU4M_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 5) | NC score | 0.521423 (rank : 6) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03905, Q6RL39, Q6RQN9, Q8HNR8 | Gene names | MT-ND4, MTND4, NADH4, ND4 | |||
|
Domain Architecture |
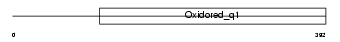
|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 4 (EC 1.6.5.3) (NADH dehydrogenase subunit 4). | |||||
|
NU2M_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 6) | NC score | 0.527815 (rank : 5) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03891, Q34769, Q9TGI0, Q9TGI1, Q9TGI2, Q9TGI3, Q9TGI4 | Gene names | MT-ND2, MTND2, NADH2, ND2 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 2 (EC 1.6.5.3) (NADH dehydrogenase subunit 2). | |||||
|
COX3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 7) | NC score | 0.033921 (rank : 7) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P00414 | Gene names | MT-CO3, COIII, COXIII, MTCO3 | |||
|
Domain Architecture |
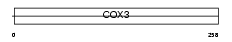
|
|||||
| Description | Cytochrome c oxidase subunit 3 (EC 1.9.3.1) (Cytochrome c oxidase polypeptide III). | |||||
|
NALP4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 8) | NC score | 0.013112 (rank : 11) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96MN2, Q86W87, Q96AY6 | Gene names | NALP4, PAN2, PYPAF4, RNH2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 4 (PYRIN-containing APAF1-like protein 4) (PAAD and NACHT-containing protein 2) (PYRIN and NACHT- containing protein 2) (Ribonuclease inhibitor 2). | |||||
|
ADA32_MOUSE
|
||||||
| θ value | 4.03905 (rank : 9) | NC score | 0.007983 (rank : 12) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K410, Q6P901, Q8BJ80 | Gene names | Adam32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAM 32 precursor (A disintegrin and metalloproteinase domain 32). | |||||
|
OR1C1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 10) | NC score | 0.000395 (rank : 13) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15619, Q5VVD2, Q6IF97, Q8NGZ1, Q96R83 | Gene names | OR1C1 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 1C1 (Olfactory receptor TPCR27) (Olfactory receptor OR1-42). | |||||
|
TXD11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 11) | NC score | 0.016331 (rank : 8) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PKC3, O95887, Q6PJA6, Q8N2Q4, Q96K45, Q96K53 | Gene names | TXNDC11, EFP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11 (EF-hand-binding protein 1). | |||||
|
FLVC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 12) | NC score | 0.013333 (rank : 10) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91X85 | Gene names | Flvcr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Feline leukemia virus subgroup C receptor-related protein 2 (Calcium- chelate transporter) (CCT). | |||||
|
PIGU_HUMAN
|
||||||
| θ value | 8.99809 (rank : 13) | NC score | 0.015894 (rank : 9) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H490, Q7Z489, Q8N2F2 | Gene names | CDC91L1, PIGU | |||
|
Domain Architecture |

|
|||||
| Description | GPI transamidase component PIG-U (Phosphatidylinositol-glycan biosynthesis class U protein) (Cell division cycle protein 91-like 1) (CDC91-like 1 protein). | |||||
|
NU5M_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P03915, Q34773, Q8WCY3 | Gene names | MT-ND5, MTND5, NADH5, ND5 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 5 (EC 1.6.5.3) (NADH dehydrogenase subunit 5). | |||||
|
NU5M_MOUSE
|
||||||
| NC score | 0.990702 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03921 | Gene names | Mtnd5, mt-Nd5, Nd5 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 5 (EC 1.6.5.3) (NADH dehydrogenase subunit 5). | |||||
|
NU2M_MOUSE
|
||||||
| NC score | 0.551802 (rank : 3) | θ value | 2.28291e-11 (rank : 3) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03893 | Gene names | Mtnd2, mt-Nd2, Nd2 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 2 (EC 1.6.5.3) (NADH dehydrogenase subunit 2). | |||||
|
NU4M_MOUSE
|
||||||
| NC score | 0.535518 (rank : 4) | θ value | 2.98157e-11 (rank : 4) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03911 | Gene names | Mtnd4, mt-Nd4, Nd4 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 4 (EC 1.6.5.3) (NADH dehydrogenase subunit 4). | |||||
|
NU2M_HUMAN
|
||||||
| NC score | 0.527815 (rank : 5) | θ value | 4.0297e-08 (rank : 6) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03891, Q34769, Q9TGI0, Q9TGI1, Q9TGI2, Q9TGI3, Q9TGI4 | Gene names | MT-ND2, MTND2, NADH2, ND2 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 2 (EC 1.6.5.3) (NADH dehydrogenase subunit 2). | |||||
|
NU4M_HUMAN
|
||||||
| NC score | 0.521423 (rank : 6) | θ value | 1.25267e-09 (rank : 5) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03905, Q6RL39, Q6RQN9, Q8HNR8 | Gene names | MT-ND4, MTND4, NADH4, ND4 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 4 (EC 1.6.5.3) (NADH dehydrogenase subunit 4). | |||||
|
COX3_HUMAN
|
||||||
| NC score | 0.033921 (rank : 7) | θ value | 3.0926 (rank : 7) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P00414 | Gene names | MT-CO3, COIII, COXIII, MTCO3 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome c oxidase subunit 3 (EC 1.9.3.1) (Cytochrome c oxidase polypeptide III). | |||||
|
TXD11_HUMAN
|
||||||
| NC score | 0.016331 (rank : 8) | θ value | 5.27518 (rank : 11) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PKC3, O95887, Q6PJA6, Q8N2Q4, Q96K45, Q96K53 | Gene names | TXNDC11, EFP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11 (EF-hand-binding protein 1). | |||||
|
PIGU_HUMAN
|
||||||
| NC score | 0.015894 (rank : 9) | θ value | 8.99809 (rank : 13) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H490, Q7Z489, Q8N2F2 | Gene names | CDC91L1, PIGU | |||
|
Domain Architecture |

|
|||||
| Description | GPI transamidase component PIG-U (Phosphatidylinositol-glycan biosynthesis class U protein) (Cell division cycle protein 91-like 1) (CDC91-like 1 protein). | |||||
|
FLVC2_MOUSE
|
||||||
| NC score | 0.013333 (rank : 10) | θ value | 8.99809 (rank : 12) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91X85 | Gene names | Flvcr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Feline leukemia virus subgroup C receptor-related protein 2 (Calcium- chelate transporter) (CCT). | |||||
|
NALP4_HUMAN
|
||||||
| NC score | 0.013112 (rank : 11) | θ value | 3.0926 (rank : 8) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96MN2, Q86W87, Q96AY6 | Gene names | NALP4, PAN2, PYPAF4, RNH2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 4 (PYRIN-containing APAF1-like protein 4) (PAAD and NACHT-containing protein 2) (PYRIN and NACHT- containing protein 2) (Ribonuclease inhibitor 2). | |||||
|
ADA32_MOUSE
|
||||||
| NC score | 0.007983 (rank : 12) | θ value | 4.03905 (rank : 9) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K410, Q6P901, Q8BJ80 | Gene names | Adam32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAM 32 precursor (A disintegrin and metalloproteinase domain 32). | |||||
|
OR1C1_HUMAN
|
||||||
| NC score | 0.000395 (rank : 13) | θ value | 5.27518 (rank : 10) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15619, Q5VVD2, Q6IF97, Q8NGZ1, Q96R83 | Gene names | OR1C1 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 1C1 (Olfactory receptor TPCR27) (Olfactory receptor OR1-42). | |||||