Please be patient as the page loads
|
NU4M_HUMAN
|
||||||
| SwissProt Accessions | P03905, Q6RL39, Q6RQN9, Q8HNR8 | Gene names | MT-ND4, MTND4, NADH4, ND4 | |||
|
Domain Architecture |
|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 4 (EC 1.6.5.3) (NADH dehydrogenase subunit 4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NU4M_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P03905, Q6RL39, Q6RQN9, Q8HNR8 | Gene names | MT-ND4, MTND4, NADH4, ND4 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 4 (EC 1.6.5.3) (NADH dehydrogenase subunit 4). | |||||
|
NU4M_MOUSE
|
||||||
| θ value | 1.05009e-157 (rank : 2) | NC score | 0.991318 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P03911 | Gene names | Mtnd4, mt-Nd4, Nd4 | |||
|
Domain Architecture |
|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 4 (EC 1.6.5.3) (NADH dehydrogenase subunit 4). | |||||
|
NU5M_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 3) | NC score | 0.569181 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03921 | Gene names | Mtnd5, mt-Nd5, Nd5 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 5 (EC 1.6.5.3) (NADH dehydrogenase subunit 5). | |||||
|
NU5M_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 4) | NC score | 0.521423 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03915, Q34773, Q8WCY3 | Gene names | MT-ND5, MTND5, NADH5, ND5 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 5 (EC 1.6.5.3) (NADH dehydrogenase subunit 5). | |||||
|
NU2M_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 5) | NC score | 0.520140 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03893 | Gene names | Mtnd2, mt-Nd2, Nd2 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 2 (EC 1.6.5.3) (NADH dehydrogenase subunit 2). | |||||
|
NU2M_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 6) | NC score | 0.456850 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03891, Q34769, Q9TGI0, Q9TGI1, Q9TGI2, Q9TGI3, Q9TGI4 | Gene names | MT-ND2, MTND2, NADH2, ND2 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 2 (EC 1.6.5.3) (NADH dehydrogenase subunit 2). | |||||
|
PAR2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 7) | NC score | 0.012759 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 576 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55085, Q13317, Q13346 | Gene names | F2RL1, GPR11, PAR2 | |||
|
Domain Architecture |
|
|||||
| Description | Proteinase-activated receptor 2 precursor (PAR-2) (Thrombin receptor- like 1) (Coagulation factor II receptor-like 1) (G-protein coupled receptor 11). | |||||
|
YIPF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 8) | NC score | 0.041006 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y548, Q9NWJ1 | Gene names | YIPF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein YIPF1 (YIP1 family member 1). | |||||
|
TR125_MOUSE
|
||||||
| θ value | 2.36792 (rank : 9) | NC score | 0.014838 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7M710 | Gene names | Tas2r125, T2r59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 2 member 125 (T2R125) (mT2R59). | |||||
|
P2RY1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 10) | NC score | 0.009104 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P47900 | Gene names | P2RY1 | |||
|
Domain Architecture |
|
|||||
| Description | P2Y purinoceptor 1 (ATP receptor) (P2Y1) (Purinergic receptor). | |||||
|
CCKAR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 11) | NC score | 0.005694 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08786, Q8VCC7, Q9DBV6 | Gene names | Cckar | |||
|
Domain Architecture |
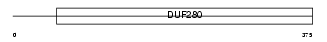
|
|||||
| Description | Cholecystokinin type A receptor (CCK-A receptor) (CCK-AR) (Cholecystokinin-1 receptor) (CCK1-R). | |||||
|
CXCR2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 12) | NC score | 0.006614 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35343 | Gene names | Il8rb, Cmkar2, Cxcr2, Gpcr16 | |||
|
Domain Architecture |
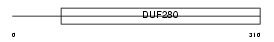
|
|||||
| Description | High affinity interleukin-8 receptor B (IL-8R B) (CXCR-2) (GRO/MGSA receptor) (CD182 antigen). | |||||
|
O2T12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 13) | NC score | 0.002151 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 656 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NG77 | Gene names | OR2T12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 2T12 (Olfactory receptor OR1-57). | |||||
|
YIPF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 14) | NC score | 0.035170 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VU1, Q3TZU1 | Gene names | Yipf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein YIPF1 (YIP1 family member 1). | |||||
|
CD1D_HUMAN
|
||||||
| θ value | 6.88961 (rank : 15) | NC score | 0.007216 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15813, Q9UMM3, Q9Y5M4 | Gene names | CD1D | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface glycoprotein CD1d precursor (CD1d antigen) (R3G1). | |||||
|
PAR2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 16) | NC score | 0.010018 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 619 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55086 | Gene names | F2rl1, Gpcr11, Gpr11, Par2 | |||
|
Domain Architecture |
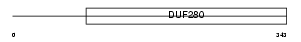
|
|||||
| Description | Proteinase-activated receptor 2 precursor (PAR-2) (Thrombin receptor- like 1) (Coagulation factor II receptor-like 1) (G-protein coupled receptor 11). | |||||
|
VN1B7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 17) | NC score | 0.006680 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQ46 | Gene names | V1rb7, V1ra11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeronasal type-1 receptor B7 (Vomeronasal type-1 receptor A11). | |||||
|
NU4M_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P03905, Q6RL39, Q6RQN9, Q8HNR8 | Gene names | MT-ND4, MTND4, NADH4, ND4 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 4 (EC 1.6.5.3) (NADH dehydrogenase subunit 4). | |||||
|
NU4M_MOUSE
|
||||||
| NC score | 0.991318 (rank : 2) | θ value | 1.05009e-157 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P03911 | Gene names | Mtnd4, mt-Nd4, Nd4 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 4 (EC 1.6.5.3) (NADH dehydrogenase subunit 4). | |||||
|
NU5M_MOUSE
|
||||||
| NC score | 0.569181 (rank : 3) | θ value | 2.20605e-14 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03921 | Gene names | Mtnd5, mt-Nd5, Nd5 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 5 (EC 1.6.5.3) (NADH dehydrogenase subunit 5). | |||||
|
NU5M_HUMAN
|
||||||
| NC score | 0.521423 (rank : 4) | θ value | 1.25267e-09 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03915, Q34773, Q8WCY3 | Gene names | MT-ND5, MTND5, NADH5, ND5 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 5 (EC 1.6.5.3) (NADH dehydrogenase subunit 5). | |||||
|
NU2M_MOUSE
|
||||||
| NC score | 0.520140 (rank : 5) | θ value | 2.13673e-09 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03893 | Gene names | Mtnd2, mt-Nd2, Nd2 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 2 (EC 1.6.5.3) (NADH dehydrogenase subunit 2). | |||||
|
NU2M_HUMAN
|
||||||
| NC score | 0.456850 (rank : 6) | θ value | 1.87187e-05 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03891, Q34769, Q9TGI0, Q9TGI1, Q9TGI2, Q9TGI3, Q9TGI4 | Gene names | MT-ND2, MTND2, NADH2, ND2 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 2 (EC 1.6.5.3) (NADH dehydrogenase subunit 2). | |||||
|
YIPF1_HUMAN
|
||||||
| NC score | 0.041006 (rank : 7) | θ value | 1.81305 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y548, Q9NWJ1 | Gene names | YIPF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein YIPF1 (YIP1 family member 1). | |||||
|
YIPF1_MOUSE
|
||||||
| NC score | 0.035170 (rank : 8) | θ value | 5.27518 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VU1, Q3TZU1 | Gene names | Yipf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein YIPF1 (YIP1 family member 1). | |||||
|
TR125_MOUSE
|
||||||
| NC score | 0.014838 (rank : 9) | θ value | 2.36792 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7M710 | Gene names | Tas2r125, T2r59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 2 member 125 (T2R125) (mT2R59). | |||||
|
PAR2_HUMAN
|
||||||
| NC score | 0.012759 (rank : 10) | θ value | 1.06291 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 576 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55085, Q13317, Q13346 | Gene names | F2RL1, GPR11, PAR2 | |||
|
Domain Architecture |

|
|||||
| Description | Proteinase-activated receptor 2 precursor (PAR-2) (Thrombin receptor- like 1) (Coagulation factor II receptor-like 1) (G-protein coupled receptor 11). | |||||
|
PAR2_MOUSE
|
||||||
| NC score | 0.010018 (rank : 11) | θ value | 6.88961 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 619 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55086 | Gene names | F2rl1, Gpcr11, Gpr11, Par2 | |||
|
Domain Architecture |

|
|||||
| Description | Proteinase-activated receptor 2 precursor (PAR-2) (Thrombin receptor- like 1) (Coagulation factor II receptor-like 1) (G-protein coupled receptor 11). | |||||
|
P2RY1_HUMAN
|
||||||
| NC score | 0.009104 (rank : 12) | θ value | 4.03905 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P47900 | Gene names | P2RY1 | |||
|
Domain Architecture |

|
|||||
| Description | P2Y purinoceptor 1 (ATP receptor) (P2Y1) (Purinergic receptor). | |||||
|
CD1D_HUMAN
|
||||||
| NC score | 0.007216 (rank : 13) | θ value | 6.88961 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15813, Q9UMM3, Q9Y5M4 | Gene names | CD1D | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface glycoprotein CD1d precursor (CD1d antigen) (R3G1). | |||||
|
VN1B7_MOUSE
|
||||||
| NC score | 0.006680 (rank : 14) | θ value | 8.99809 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQ46 | Gene names | V1rb7, V1ra11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeronasal type-1 receptor B7 (Vomeronasal type-1 receptor A11). | |||||
|
CXCR2_MOUSE
|
||||||
| NC score | 0.006614 (rank : 15) | θ value | 5.27518 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35343 | Gene names | Il8rb, Cmkar2, Cxcr2, Gpcr16 | |||
|
Domain Architecture |

|
|||||
| Description | High affinity interleukin-8 receptor B (IL-8R B) (CXCR-2) (GRO/MGSA receptor) (CD182 antigen). | |||||
|
CCKAR_MOUSE
|
||||||
| NC score | 0.005694 (rank : 16) | θ value | 5.27518 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08786, Q8VCC7, Q9DBV6 | Gene names | Cckar | |||
|
Domain Architecture |

|
|||||
| Description | Cholecystokinin type A receptor (CCK-A receptor) (CCK-AR) (Cholecystokinin-1 receptor) (CCK1-R). | |||||
|
O2T12_HUMAN
|
||||||
| NC score | 0.002151 (rank : 17) | θ value | 5.27518 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 656 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NG77 | Gene names | OR2T12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 2T12 (Olfactory receptor OR1-57). | |||||