Please be patient as the page loads
|
ASAH3_MOUSE
|
||||||
| SwissProt Accessions | Q8R4X1 | Gene names | Asah3, Acer1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkaline ceramidase 1 (EC 3.5.1.23) (Alkaline CDase-1) (AlkCDase 1) (maCER1) (Acylsphingosine deacylase 3) (N-acylsphingosine amidohydrolase 3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ASAH3_MOUSE
|
||||||
| θ value | 1.7349e-152 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R4X1 | Gene names | Asah3, Acer1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkaline ceramidase 1 (EC 3.5.1.23) (Alkaline CDase-1) (AlkCDase 1) (maCER1) (Acylsphingosine deacylase 3) (N-acylsphingosine amidohydrolase 3). | |||||
|
ASAH3_HUMAN
|
||||||
| θ value | 5.26011e-125 (rank : 2) | NC score | 0.996927 (rank : 2) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TDN7 | Gene names | ASAH3, ACER1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkaline ceramidase 1 (EC 3.5.1.23) (Alkaline CDase-1) (AlkCDase 1) (Acylsphingosine deacylase 3) (N-acylsphingosine amidohydrolase 3). | |||||
|
ASA3L_MOUSE
|
||||||
| θ value | 5.93839e-52 (rank : 3) | NC score | 0.929649 (rank : 3) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VD53, Q6PB92, Q8BUG3 | Gene names | Asah3l, Acer2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkaline ceramidase 2 (EC 3.5.1.23) (AlkCDase 2) (maCER2) (Acylsphingosine deacylase 3-like) (N-acylsphingosine amidohydrolase 3-like) (Cancer related gene-liver 1) (CRG-L1). | |||||
|
ASA3L_HUMAN
|
||||||
| θ value | 7.75577e-52 (rank : 4) | NC score | 0.929263 (rank : 4) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5QJU3, Q569G5, Q5VZR7, Q71RD2 | Gene names | ASAH3L, ACER2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkaline ceramidase 2 (EC 3.5.1.23) (AlkCDase 2) (Acylsphingosine deacylase 3-like) (N-acylsphingosine amidohydrolase 3-like). | |||||
|
APHC_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 5) | NC score | 0.597307 (rank : 5) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D099, Q542R2, Q9D0X4, Q9D3J4 | Gene names | Phca, Aphc | |||
|
Domain Architecture |
|
|||||
| Description | Alkaline phytoceramidase (EC 3.5.1.-) (aPHC) (Alkaline ceramidase). | |||||
|
APHC_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 6) | NC score | 0.588451 (rank : 6) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NUN7 | Gene names | PHCA, APHC | |||
|
Domain Architecture |
|
|||||
| Description | Alkaline phytoceramidase (EC 3.5.1.-) (aPHC) (Alkaline ceramidase) (Alkaline dihydroceramidase SB89). | |||||
|
TRPC2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 7) | NC score | 0.027967 (rank : 11) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R244, Q9ES59, Q9ES60, Q9R243 | Gene names | Trpc2, Trp2, Trrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 2 (TrpC2) (mTrp2). | |||||
|
TR110_MOUSE
|
||||||
| θ value | 1.06291 (rank : 8) | NC score | 0.032134 (rank : 10) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7M712, Q71QD1 | Gene names | Tas2r110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 2 member 110 (T2R110) (mT2r57) (STC 9-1). | |||||
|
TR123_MOUSE
|
||||||
| θ value | 1.06291 (rank : 9) | NC score | 0.032293 (rank : 9) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59528, Q7M714 | Gene names | Tas2r123, T2r55, Tas2r23 | |||
|
Domain Architecture |
|
|||||
| Description | Taste receptor type 2 member 123 (T2R123) (Taste receptor type 2 member 23) (T2R23) (mT2R55) (STC9-2). | |||||
|
PORCN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 10) | NC score | 0.066078 (rank : 7) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H237, Q14829, Q9H234, Q9H235, Q9H236, Q9UJU7 | Gene names | PORCN, MG61, PORC, PPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable protein-cysteine N-palmitoyltransferase porcupine (EC 2.3.1.-) (Protein MG61). | |||||
|
PORCN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 11) | NC score | 0.064402 (rank : 8) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJJ7, Q9CWT0, Q9JJJ8, Q9JJJ9, Q9JJK0 | Gene names | Porcn, Porc, Ppn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable protein-cysteine N-palmitoyltransferase porcupine (EC 2.3.1.-) (mPORC). | |||||
|
T2R10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 12) | NC score | 0.019891 (rank : 12) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NYW0, Q3MIM9, Q6NTD9 | Gene names | TAS2R10 | |||
|
Domain Architecture |
|
|||||
| Description | Taste receptor type 2 member 10 (T2R10) (Taste receptor family B member 2) (TRB2). | |||||
|
NU4M_MOUSE
|
||||||
| θ value | 6.88961 (rank : 13) | NC score | 0.017641 (rank : 13) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P03911 | Gene names | Mtnd4, mt-Nd4, Nd4 | |||
|
Domain Architecture |
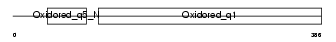
|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 4 (EC 1.6.5.3) (NADH dehydrogenase subunit 4). | |||||
|
ASAH3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.7349e-152 (rank : 1) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R4X1 | Gene names | Asah3, Acer1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkaline ceramidase 1 (EC 3.5.1.23) (Alkaline CDase-1) (AlkCDase 1) (maCER1) (Acylsphingosine deacylase 3) (N-acylsphingosine amidohydrolase 3). | |||||
|
ASAH3_HUMAN
|
||||||
| NC score | 0.996927 (rank : 2) | θ value | 5.26011e-125 (rank : 2) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TDN7 | Gene names | ASAH3, ACER1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkaline ceramidase 1 (EC 3.5.1.23) (Alkaline CDase-1) (AlkCDase 1) (Acylsphingosine deacylase 3) (N-acylsphingosine amidohydrolase 3). | |||||
|
ASA3L_MOUSE
|
||||||
| NC score | 0.929649 (rank : 3) | θ value | 5.93839e-52 (rank : 3) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VD53, Q6PB92, Q8BUG3 | Gene names | Asah3l, Acer2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkaline ceramidase 2 (EC 3.5.1.23) (AlkCDase 2) (maCER2) (Acylsphingosine deacylase 3-like) (N-acylsphingosine amidohydrolase 3-like) (Cancer related gene-liver 1) (CRG-L1). | |||||
|
ASA3L_HUMAN
|
||||||
| NC score | 0.929263 (rank : 4) | θ value | 7.75577e-52 (rank : 4) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5QJU3, Q569G5, Q5VZR7, Q71RD2 | Gene names | ASAH3L, ACER2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkaline ceramidase 2 (EC 3.5.1.23) (AlkCDase 2) (Acylsphingosine deacylase 3-like) (N-acylsphingosine amidohydrolase 3-like). | |||||
|
APHC_MOUSE
|
||||||
| NC score | 0.597307 (rank : 5) | θ value | 3.89403e-11 (rank : 5) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D099, Q542R2, Q9D0X4, Q9D3J4 | Gene names | Phca, Aphc | |||
|
Domain Architecture |

|
|||||
| Description | Alkaline phytoceramidase (EC 3.5.1.-) (aPHC) (Alkaline ceramidase). | |||||
|
APHC_HUMAN
|
||||||
| NC score | 0.588451 (rank : 6) | θ value | 5.62301e-10 (rank : 6) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NUN7 | Gene names | PHCA, APHC | |||
|
Domain Architecture |

|
|||||
| Description | Alkaline phytoceramidase (EC 3.5.1.-) (aPHC) (Alkaline ceramidase) (Alkaline dihydroceramidase SB89). | |||||
|
PORCN_HUMAN
|
||||||
| NC score | 0.066078 (rank : 7) | θ value | 1.81305 (rank : 10) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H237, Q14829, Q9H234, Q9H235, Q9H236, Q9UJU7 | Gene names | PORCN, MG61, PORC, PPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable protein-cysteine N-palmitoyltransferase porcupine (EC 2.3.1.-) (Protein MG61). | |||||
|
PORCN_MOUSE
|
||||||
| NC score | 0.064402 (rank : 8) | θ value | 2.36792 (rank : 11) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJJ7, Q9CWT0, Q9JJJ8, Q9JJJ9, Q9JJK0 | Gene names | Porcn, Porc, Ppn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable protein-cysteine N-palmitoyltransferase porcupine (EC 2.3.1.-) (mPORC). | |||||
|
TR123_MOUSE
|
||||||
| NC score | 0.032293 (rank : 9) | θ value | 1.06291 (rank : 9) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59528, Q7M714 | Gene names | Tas2r123, T2r55, Tas2r23 | |||
|
Domain Architecture |

|
|||||
| Description | Taste receptor type 2 member 123 (T2R123) (Taste receptor type 2 member 23) (T2R23) (mT2R55) (STC9-2). | |||||
|
TR110_MOUSE
|
||||||
| NC score | 0.032134 (rank : 10) | θ value | 1.06291 (rank : 8) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7M712, Q71QD1 | Gene names | Tas2r110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 2 member 110 (T2R110) (mT2r57) (STC 9-1). | |||||
|
TRPC2_MOUSE
|
||||||
| NC score | 0.027967 (rank : 11) | θ value | 0.279714 (rank : 7) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R244, Q9ES59, Q9ES60, Q9R243 | Gene names | Trpc2, Trp2, Trrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 2 (TrpC2) (mTrp2). | |||||
|
T2R10_HUMAN
|
||||||
| NC score | 0.019891 (rank : 12) | θ value | 4.03905 (rank : 12) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NYW0, Q3MIM9, Q6NTD9 | Gene names | TAS2R10 | |||
|
Domain Architecture |

|
|||||
| Description | Taste receptor type 2 member 10 (T2R10) (Taste receptor family B member 2) (TRB2). | |||||
|
NU4M_MOUSE
|
||||||
| NC score | 0.017641 (rank : 13) | θ value | 6.88961 (rank : 13) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P03911 | Gene names | Mtnd4, mt-Nd4, Nd4 | |||
|
Domain Architecture |

|
|||||
| Description | NADH-ubiquinone oxidoreductase chain 4 (EC 1.6.5.3) (NADH dehydrogenase subunit 4). | |||||