Please be patient as the page loads
|
TRPC2_MOUSE
|
||||||
| SwissProt Accessions | Q9R244, Q9ES59, Q9ES60, Q9R243 | Gene names | Trpc2, Trp2, Trrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 2 (TrpC2) (mTrp2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TRPC2_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9R244, Q9ES59, Q9ES60, Q9R243 | Gene names | Trpc2, Trp2, Trrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 2 (TrpC2) (mTrp2). | |||||
|
TRPC3_HUMAN
|
||||||
| θ value | 6.86991e-125 (rank : 2) | NC score | 0.946199 (rank : 9) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13507, O00593, Q15660 | Gene names | TRPC3, TRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Htrp-3) (Htrp3). | |||||
|
TRPC3_MOUSE
|
||||||
| θ value | 9.92011e-124 (rank : 3) | NC score | 0.948796 (rank : 2) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QZC1, Q61058 | Gene names | Trpc3, Trp3, Trrp3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Transient receptor protein 3) (Mtrp3) (Receptor-activated cation channel TRP3) (trp-related protein 3). | |||||
|
TRPC6_HUMAN
|
||||||
| θ value | 3.52827e-121 (rank : 4) | NC score | 0.942432 (rank : 11) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y210, Q9HCW3, Q9NQA8, Q9NQA9 | Gene names | TRPC6, TRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6). | |||||
|
TRPC6_MOUSE
|
||||||
| θ value | 5.63297e-119 (rank : 5) | NC score | 0.946115 (rank : 10) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
TRPC7_HUMAN
|
||||||
| θ value | 9.30652e-114 (rank : 6) | NC score | 0.947118 (rank : 8) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HCX4 | Gene names | TRPC7, TRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (TRP7 protein). | |||||
|
TRPC7_MOUSE
|
||||||
| θ value | 4.61877e-113 (rank : 7) | NC score | 0.947627 (rank : 7) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9WVC5 | Gene names | Trpc7, Trp7, Trrp8 | |||
|
Domain Architecture |
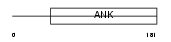
|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (mTRP7). | |||||
|
TRPC4_MOUSE
|
||||||
| θ value | 4.04623e-109 (rank : 8) | NC score | 0.947942 (rank : 4) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QUQ5, Q62350, Q9QUQ9, Q9QZC0 | Gene names | Trpc4, Trrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (Receptor- activated cation channel TRP4) (Capacitative calcium entry channel Trp4). | |||||
|
TRPC4_HUMAN
|
||||||
| θ value | 1.53757e-108 (rank : 9) | NC score | 0.948122 (rank : 3) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
|
TRPC5_MOUSE
|
||||||
| θ value | 3.10529e-101 (rank : 10) | NC score | 0.947849 (rank : 5) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QX29, Q61059, Q9QWT1, Q9R0D4 | Gene names | Trpc5, Trp5, Trrp5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Transient receptor protein 5) (Mtrp5) (trp-related protein 5) (Capacitative calcium entry channel 2) (CCE2). | |||||
|
TRPC5_HUMAN
|
||||||
| θ value | 1.18001e-100 (rank : 11) | NC score | 0.947630 (rank : 6) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UL62, O75233, Q5JXY8, Q9Y514 | Gene names | TRPC5, TRP5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Htrp-5) (Htrp5). | |||||
|
TRPC1_HUMAN
|
||||||
| θ value | 3.0147e-88 (rank : 12) | NC score | 0.940615 (rank : 13) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P48995 | Gene names | TRPC1, TRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (TRP-1 protein). | |||||
|
TRPC1_MOUSE
|
||||||
| θ value | 3.0147e-88 (rank : 13) | NC score | 0.941288 (rank : 12) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61056, O35722 | Gene names | Trpc1, Trp1, Trrp1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (Transient receptor protein 1) (Mtrp1) (Trp-related protein 1). | |||||
|
TRPM8_MOUSE
|
||||||
| θ value | 8.38298e-14 (rank : 14) | NC score | 0.530121 (rank : 14) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8R4D5 | Gene names | Trpm8, Trpp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8. | |||||
|
TRPM8_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 15) | NC score | 0.526969 (rank : 15) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7Z2W7, Q6QNH9, Q8TAC3, Q8TDX8, Q9BVK1 | Gene names | TRPM8, LTRPC6, TRPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8 (Transient receptor potential-p8) (Trp-p8) (Long transient receptor potential channel 6) (LTrpC6). | |||||
|
TRPM7_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 16) | NC score | 0.441360 (rank : 17) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
TRPM7_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 17) | NC score | 0.444039 (rank : 16) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
TRPM6_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 18) | NC score | 0.435137 (rank : 19) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CIR4 | Gene names | Trpm6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM6_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 19) | NC score | 0.412858 (rank : 21) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BX84, Q6VPR8, Q6VPR9, Q6VPS0, Q6VPS1, Q6VPS2 | Gene names | TRPM6, CHAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
XRCC1_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 20) | NC score | 0.134812 (rank : 37) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P18887, Q9HCB1 | Gene names | XRCC1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
TRPV6_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 21) | NC score | 0.234559 (rank : 28) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H1D0, Q8TDL3, Q8WXR8, Q96LC5, Q9H1D1, Q9H296 | Gene names | TRPV6, ECAC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1) (CaT-like) (CaT-L). | |||||
|
TRPV6_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 22) | NC score | 0.220633 (rank : 30) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91WD2 | Gene names | Trpv6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1). | |||||
|
XRCC1_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 23) | NC score | 0.122541 (rank : 38) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
TRPV5_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 24) | NC score | 0.237084 (rank : 27) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P69744 | Gene names | Trpv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
TRPM3_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 25) | NC score | 0.363806 (rank : 22) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9HCF6, Q5VW02, Q5VW03, Q5VW04, Q5W5T7, Q86SH0, Q86SH6, Q86UL0, Q86WK1, Q86WK2, Q86WK3, Q86WK4, Q86YZ9, Q86Z00, Q86Z01 | Gene names | TRPM3, KIAA1616, LTRPC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 3 (Long transient receptor potential channel 3) (LTrpC3) (Melastatin-2) (MLSN2). | |||||
|
TRPV3_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 26) | NC score | 0.313938 (rank : 23) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
TRPM2_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 27) | NC score | 0.414491 (rank : 20) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91YD4 | Gene names | Trpm2, Ltrpc2, Trpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7). | |||||
|
TRPV5_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 28) | NC score | 0.206211 (rank : 32) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
TRPV3_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 29) | NC score | 0.293945 (rank : 24) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
TRPV1_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 30) | NC score | 0.261063 (rank : 26) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q704Y3, Q5SSE1, Q5SSE2, Q5SSE4, Q5WPV5, Q68SW0 | Gene names | Trpv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1). | |||||
|
TRPM2_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 31) | NC score | 0.438888 (rank : 18) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O94759, Q5KTC2, Q6J3P5, Q96KN6, Q96Q93 | Gene names | TRPM2, EREG1, KNP3, LTRPC2, TRPC7 | |||
|
Domain Architecture |

|
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7) (Estrogen- responsive element-associated gene 1 protein). | |||||
|
TRPV1_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 32) | NC score | 0.261385 (rank : 25) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8NER1, Q9H0G9, Q9H303, Q9H304, Q9NQ74, Q9NY22 | Gene names | TRPV1, VR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1) (Capsaicin receptor). | |||||
|
RN126_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 33) | NC score | 0.042255 (rank : 52) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BV68, Q9NWX1 | Gene names | RNF126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 126. | |||||
|
RN126_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 34) | NC score | 0.041325 (rank : 53) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YL2 | Gene names | Rnf126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 126. | |||||
|
TATD2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 35) | NC score | 0.054302 (rank : 47) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q93075 | Gene names | TATDN2, KIAA0218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TatD DNase domain-containing deoxyribonuclease 2 (EC 3.1.21.-). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 36) | NC score | 0.017093 (rank : 65) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
TRPV4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 37) | NC score | 0.227115 (rank : 29) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
ANR25_MOUSE
|
||||||
| θ value | 0.279714 (rank : 38) | NC score | 0.029729 (rank : 56) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BX02, Q8BLD5, Q91Z35 | Gene names | Ankrd25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25. | |||||
|
ASAH3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 39) | NC score | 0.027967 (rank : 57) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R4X1 | Gene names | Asah3, Acer1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkaline ceramidase 1 (EC 3.5.1.23) (Alkaline CDase-1) (AlkCDase 1) (maCER1) (Acylsphingosine deacylase 3) (N-acylsphingosine amidohydrolase 3). | |||||
|
TRPV4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 40) | NC score | 0.210318 (rank : 31) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9EPK8, Q91XR5, Q9EQZ4, Q9ERZ7, Q9ES76 | Gene names | Trpv4, Trp12, Vrl2, Vroac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (Vanilloid receptor- related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 41) | NC score | 0.029904 (rank : 55) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 0.365318 (rank : 42) | NC score | 0.021812 (rank : 60) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.045438 (rank : 51) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
ZFP95_HUMAN
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | -0.002212 (rank : 96) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2L8 | Gene names | ZFP95, KIAA1015 | |||
|
Domain Architecture |
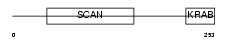
|
|||||
| Description | Zinc finger protein 95 homolog (Zfp-95). | |||||
|
K0562_MOUSE
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.020036 (rank : 63) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80V31, Q6ZQ95 | Gene names | Kiaa0562 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0562. | |||||
|
KSR1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.000152 (rank : 94) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61097, Q61648, Q78DX8 | Gene names | Ksr1, Ksr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras) (mKSR1) (Hb protein). | |||||
|
NPAS4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.020929 (rank : 61) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
SC11A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.024979 (rank : 58) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.024140 (rank : 59) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
NELF_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.018905 (rank : 64) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99NF2, Q8BPT0, Q8C9R5, Q9DBF4, Q9ERZ1 | Gene names | Nelf, Jac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nasal embryonic luteinizing hormone-releasing hormone factor (Nasal embryonic LHRH factor) (Juxtasynaptic attractor of caldendrin on dendritic boutons protein) (Jacob protein). | |||||
|
NPAS4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.020175 (rank : 62) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.036418 (rank : 54) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ENAM_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.017083 (rank : 66) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
CAN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.007308 (rank : 80) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08529, O35518, O54843 | Gene names | Capn2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (80 kDa M-calpain subunit) (CALP80). | |||||
|
PAX6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.004222 (rank : 89) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26367, Q6N006, Q99413 | Gene names | PAX6, AN2 | |||
|
Domain Architecture |
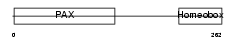
|
|||||
| Description | Paired box protein Pax-6 (Oculorhombin) (Aniridia type II protein). | |||||
|
PAX6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.004222 (rank : 90) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P63015, P32117, P70601, Q62222, Q64037, Q8CEI5, Q8VDB5, Q921Q8 | Gene names | Pax6, Pax-6, Sey | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paired box protein Pax-6 (Oculorhombin). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.006780 (rank : 81) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
C2TA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.010259 (rank : 71) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
GA2L2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.009039 (rank : 76) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
HRSL5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.010830 (rank : 69) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CPX5, Q9CVQ2, Q9CVS0 | Gene names | Hrasls5, Hrlp5 | |||
|
Domain Architecture |

|
|||||
| Description | HRAS-like suppressor 5 (H-rev107-like protein 5). | |||||
|
MA1A1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.007322 (rank : 79) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P45700 | Gene names | Man1a1, Man1a | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IA) (Alpha-1,2-mannosidase IA) (Mannosidase alpha class 1A member 1) (Man(9)-alpha-mannosidase) (Man9-mannosidase). | |||||
|
REEP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.008298 (rank : 77) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCD6 | Gene names | Reep2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor expression-enhancing protein 2. | |||||
|
SETX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.009450 (rank : 75) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
TRPA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.060441 (rank : 46) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75762 | Gene names | TRPA1, ANKTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1) (Transformation sensitive-protein p120). | |||||
|
UBE3C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.005751 (rank : 84) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
CAN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.006606 (rank : 83) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17655, Q16738, Q6PJT3, Q8WU26, Q9HBB1 | Gene names | CAPN2, CANPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (Calpain large polypeptide L2). | |||||
|
CYLD_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.010279 (rank : 70) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80TQ2, Q80VB3, Q8BXZ3, Q8BYL9, Q8CGB0 | Gene names | Cyld, Cyld1, Kiaa0849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase CYLD (EC 3.1.2.15) (Ubiquitin thioesterase CYLD) (Ubiquitin-specific-processing protease CYLD) (Deubiquitinating enzyme CYLD). | |||||
|
ALEX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.009634 (rank : 73) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
CABP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.003272 (rank : 91) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CN133_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.009987 (rank : 72) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BGQ1, Q8BL55, Q8CIK4, Q91YI3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf133 homolog. | |||||
|
FBX21_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.009569 (rank : 74) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VDH1, Q8BHA5, Q8CHC6 | Gene names | Fbxo21, Fbx21, Kiaa0875 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 21. | |||||
|
LRC47_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.002373 (rank : 93) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q505F5, Q3U380, Q6ZPW2, Q9CUZ0 | Gene names | Lrrc47, Kiaa1185 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 47. | |||||
|
LSD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.005085 (rank : 87) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.013295 (rank : 68) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
RN5A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.045907 (rank : 50) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q05921, Q9ERU7 | Gene names | Rnasel, Rns4 | |||
|
Domain Architecture |

|
|||||
| Description | 2-5A-dependent ribonuclease (EC 3.1.26.-) (2-5A-dependent RNase) (Ribonuclease L) (RNase L) (Ribonuclease 4). | |||||
|
ZN672_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | -0.003277 (rank : 97) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99LH4, Q8BIR7 | Gene names | Znf672, Zfp672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 672. | |||||
|
DGAT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.006750 (rank : 82) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2A7, Q9D7Q5 | Gene names | Dgat1, Dgat | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol O-acyltransferase 1 (EC 2.3.1.20) (Diglyceride acyltransferase). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.005230 (rank : 86) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
MFN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.005279 (rank : 85) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95140, O95572, Q5JXC3, Q5JXC4, Q9H131, Q9NSX8 | Gene names | MFN2, CPRP1, KIAA0214 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane GTPase MFN2 (EC 3.6.5.-) (Mitofusin-2). | |||||
|
PSD10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.014041 (rank : 67) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75832 | Gene names | PSMD10 | |||
|
Domain Architecture |
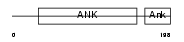
|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
RBBP8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.008110 (rank : 78) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99708, O75371 | Gene names | RBBP8, CTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 8 (RBBP-8) (CtBP-interacting protein) (CtIP) (Retinoblastoma-interacting protein and myosin-like) (RIM). | |||||
|
RIMS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.004426 (rank : 88) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
ROBO3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | -0.001079 (rank : 95) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96MS0 | Gene names | ROBO3 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Roundabout-like protein 3). | |||||
|
TTF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.002673 (rank : 92) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
ACBD6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.065848 (rank : 44) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D061, Q3TMN7, Q9DCU4 | Gene names | Acbd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyl-CoA-binding domain-containing protein 6. | |||||
|
ANK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.067419 (rank : 42) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
ANK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.067379 (rank : 43) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
ANKS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.053716 (rank : 48) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92625, Q5JYI9, Q5SYR2, Q86WQ7 | Gene names | ANKS1A, ANKS1, KIAA0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A (Odin). | |||||
|
ANKY2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.065821 (rank : 45) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3TPE9, Q8BK14, Q8BYW5, Q8R3N4, Q921J1 | Gene names | Ankmy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
PK2L1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.168088 (rank : 33) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PK2L2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.099821 (rank : 41) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.137708 (rank : 36) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PKD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.120491 (rank : 39) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
PKD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.118911 (rank : 40) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
TRPA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.050383 (rank : 49) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BLA8 | Gene names | Trpa1, Anktm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1). | |||||
|
TRPV2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.159166 (rank : 35) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y5S1, Q9Y670 | Gene names | TRPV2, VRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Vanilloid receptor-like protein 1) (VRL-1). | |||||
|
TRPV2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.166624 (rank : 34) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WTR1, Q99K71 | Gene names | Trpv2, Grc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Growth factor-regulated calcium channel) (GRC). | |||||
|
TRPC2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9R244, Q9ES59, Q9ES60, Q9R243 | Gene names | Trpc2, Trp2, Trrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 2 (TrpC2) (mTrp2). | |||||
|
TRPC3_MOUSE
|
||||||
| NC score | 0.948796 (rank : 2) | θ value | 9.92011e-124 (rank : 3) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QZC1, Q61058 | Gene names | Trpc3, Trp3, Trrp3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Transient receptor protein 3) (Mtrp3) (Receptor-activated cation channel TRP3) (trp-related protein 3). | |||||
|
TRPC4_HUMAN
|
||||||
| NC score | 0.948122 (rank : 3) | θ value | 1.53757e-108 (rank : 9) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
|
TRPC4_MOUSE
|
||||||
| NC score | 0.947942 (rank : 4) | θ value | 4.04623e-109 (rank : 8) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QUQ5, Q62350, Q9QUQ9, Q9QZC0 | Gene names | Trpc4, Trrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (Receptor- activated cation channel TRP4) (Capacitative calcium entry channel Trp4). | |||||
|
TRPC5_MOUSE
|
||||||
| NC score | 0.947849 (rank : 5) | θ value | 3.10529e-101 (rank : 10) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QX29, Q61059, Q9QWT1, Q9R0D4 | Gene names | Trpc5, Trp5, Trrp5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Transient receptor protein 5) (Mtrp5) (trp-related protein 5) (Capacitative calcium entry channel 2) (CCE2). | |||||
|
TRPC5_HUMAN
|
||||||
| NC score | 0.947630 (rank : 6) | θ value | 1.18001e-100 (rank : 11) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UL62, O75233, Q5JXY8, Q9Y514 | Gene names | TRPC5, TRP5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Htrp-5) (Htrp5). | |||||
|
TRPC7_MOUSE
|
||||||
| NC score | 0.947627 (rank : 7) | θ value | 4.61877e-113 (rank : 7) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9WVC5 | Gene names | Trpc7, Trp7, Trrp8 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (mTRP7). | |||||
|
TRPC7_HUMAN
|
||||||
| NC score | 0.947118 (rank : 8) | θ value | 9.30652e-114 (rank : 6) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HCX4 | Gene names | TRPC7, TRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (TRP7 protein). | |||||
|
TRPC3_HUMAN
|
||||||
| NC score | 0.946199 (rank : 9) | θ value | 6.86991e-125 (rank : 2) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13507, O00593, Q15660 | Gene names | TRPC3, TRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Htrp-3) (Htrp3). | |||||
|
TRPC6_MOUSE
|
||||||
| NC score | 0.946115 (rank : 10) | θ value | 5.63297e-119 (rank : 5) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
TRPC6_HUMAN
|
||||||
| NC score | 0.942432 (rank : 11) | θ value | 3.52827e-121 (rank : 4) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y210, Q9HCW3, Q9NQA8, Q9NQA9 | Gene names | TRPC6, TRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6). | |||||
|
TRPC1_MOUSE
|
||||||
| NC score | 0.941288 (rank : 12) | θ value | 3.0147e-88 (rank : 13) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61056, O35722 | Gene names | Trpc1, Trp1, Trrp1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (Transient receptor protein 1) (Mtrp1) (Trp-related protein 1). | |||||
|
TRPC1_HUMAN
|
||||||
| NC score | 0.940615 (rank : 13) | θ value | 3.0147e-88 (rank : 12) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P48995 | Gene names | TRPC1, TRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (TRP-1 protein). | |||||
|
TRPM8_MOUSE
|
||||||
| NC score | 0.530121 (rank : 14) | θ value | 8.38298e-14 (rank : 14) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8R4D5 | Gene names | Trpm8, Trpp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8. | |||||
|
TRPM8_HUMAN
|
||||||
| NC score | 0.526969 (rank : 15) | θ value | 4.16044e-13 (rank : 15) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7Z2W7, Q6QNH9, Q8TAC3, Q8TDX8, Q9BVK1 | Gene names | TRPM8, LTRPC6, TRPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8 (Transient receptor potential-p8) (Trp-p8) (Long transient receptor potential channel 6) (LTrpC6). | |||||
|
TRPM7_MOUSE
|
||||||
| NC score | 0.444039 (rank : 16) | θ value | 3.29651e-10 (rank : 17) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
TRPM7_HUMAN
|
||||||
| NC score | 0.441360 (rank : 17) | θ value | 6.64225e-11 (rank : 16) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
TRPM2_HUMAN
|
||||||
| NC score | 0.438888 (rank : 18) | θ value | 0.000461057 (rank : 31) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O94759, Q5KTC2, Q6J3P5, Q96KN6, Q96Q93 | Gene names | TRPM2, EREG1, KNP3, LTRPC2, TRPC7 | |||
|
Domain Architecture |

|
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7) (Estrogen- responsive element-associated gene 1 protein). | |||||
|
TRPM6_MOUSE
|
||||||
| NC score | 0.435137 (rank : 19) | θ value | 2.36244e-08 (rank : 18) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CIR4 | Gene names | Trpm6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM2_MOUSE
|
||||||
| NC score | 0.414491 (rank : 20) | θ value | 7.1131e-05 (rank : 27) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91YD4 | Gene names | Trpm2, Ltrpc2, Trpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7). | |||||
|
TRPM6_HUMAN
|
||||||
| NC score | 0.412858 (rank : 21) | θ value | 3.08544e-08 (rank : 19) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BX84, Q6VPR8, Q6VPR9, Q6VPS0, Q6VPS1, Q6VPS2 | Gene names | TRPM6, CHAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM3_HUMAN
|
||||||
| NC score | 0.363806 (rank : 22) | θ value | 8.40245e-06 (rank : 25) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9HCF6, Q5VW02, Q5VW03, Q5VW04, Q5W5T7, Q86SH0, Q86SH6, Q86UL0, Q86WK1, Q86WK2, Q86WK3, Q86WK4, Q86YZ9, Q86Z00, Q86Z01 | Gene names | TRPM3, KIAA1616, LTRPC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 3 (Long transient receptor potential channel 3) (LTrpC3) (Melastatin-2) (MLSN2). | |||||
|
TRPV3_HUMAN
|
||||||
| NC score | 0.313938 (rank : 23) | θ value | 1.09739e-05 (rank : 26) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
TRPV3_MOUSE
|
||||||
| NC score | 0.293945 (rank : 24) | θ value | 0.00020696 (rank : 29) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
TRPV1_HUMAN
|
||||||
| NC score | 0.261385 (rank : 25) | θ value | 0.000786445 (rank : 32) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8NER1, Q9H0G9, Q9H303, Q9H304, Q9NQ74, Q9NY22 | Gene names | TRPV1, VR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1) (Capsaicin receptor). | |||||
|
TRPV1_MOUSE
|
||||||
| NC score | 0.261063 (rank : 26) | θ value | 0.00035302 (rank : 30) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q704Y3, Q5SSE1, Q5SSE2, Q5SSE4, Q5WPV5, Q68SW0 | Gene names | Trpv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1). | |||||
|
TRPV5_MOUSE
|
||||||
| NC score | 0.237084 (rank : 27) | θ value | 1.69304e-06 (rank : 24) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P69744 | Gene names | Trpv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
TRPV6_HUMAN
|
||||||
| NC score | 0.234559 (rank : 28) | θ value | 4.45536e-07 (rank : 21) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H1D0, Q8TDL3, Q8WXR8, Q96LC5, Q9H1D1, Q9H296 | Gene names | TRPV6, ECAC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1) (CaT-like) (CaT-L). | |||||
|
TRPV4_HUMAN
|
||||||
| NC score | 0.227115 (rank : 29) | θ value | 0.163984 (rank : 37) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPV6_MOUSE
|
||||||
| NC score | 0.220633 (rank : 30) | θ value | 7.59969e-07 (rank : 22) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91WD2 | Gene names | Trpv6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1). | |||||
|
TRPV4_MOUSE
|
||||||
| NC score | 0.210318 (rank : 31) | θ value | 0.279714 (rank : 40) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9EPK8, Q91XR5, Q9EQZ4, Q9ERZ7, Q9ES76 | Gene names | Trpv4, Trp12, Vrl2, Vroac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (Vanilloid receptor- related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPV5_HUMAN
|
||||||
| NC score | 0.206211 (rank : 32) | θ value | 7.1131e-05 (rank : 28) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
PK2L1_HUMAN
|
||||||
| NC score | 0.168088 (rank : 33) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
TRPV2_MOUSE
|
||||||
| NC score | 0.166624 (rank : 34) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WTR1, Q99K71 | Gene names | Trpv2, Grc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Growth factor-regulated calcium channel) (GRC). | |||||
|
TRPV2_HUMAN
|
||||||
| NC score | 0.159166 (rank : 35) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y5S1, Q9Y670 | Gene names | TRPV2, VRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Vanilloid receptor-like protein 1) (VRL-1). | |||||
|
PK2L2_MOUSE
|
||||||
| NC score | 0.137708 (rank : 36) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
XRCC1_HUMAN
|
||||||
| NC score | 0.134812 (rank : 37) | θ value | 6.87365e-08 (rank : 20) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P18887, Q9HCB1 | Gene names | XRCC1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
XRCC1_MOUSE
|
||||||
| NC score | 0.122541 (rank : 38) | θ value | 1.29631e-06 (rank : 23) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
PKD2_HUMAN
|
||||||
| NC score | 0.120491 (rank : 39) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
PKD2_MOUSE
|
||||||
| NC score | 0.118911 (rank : 40) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
PK2L2_HUMAN
|
||||||
| NC score | 0.099821 (rank : 41) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
ANK1_HUMAN
|
||||||
| NC score | 0.067419 (rank : 42) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
ANK1_MOUSE
|
||||||
| NC score | 0.067379 (rank : 43) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
ACBD6_MOUSE
|
||||||
| NC score | 0.065848 (rank : 44) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D061, Q3TMN7, Q9DCU4 | Gene names | Acbd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyl-CoA-binding domain-containing protein 6. | |||||
|
ANKY2_MOUSE
|
||||||
| NC score | 0.065821 (rank : 45) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3TPE9, Q8BK14, Q8BYW5, Q8R3N4, Q921J1 | Gene names | Ankmy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
TRPA1_HUMAN
|
||||||
| NC score | 0.060441 (rank : 46) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75762 | Gene names | TRPA1, ANKTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1) (Transformation sensitive-protein p120). | |||||
|
TATD2_HUMAN
|
||||||
| NC score | 0.054302 (rank : 47) | θ value | 0.0431538 (rank : 35) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q93075 | Gene names | TATDN2, KIAA0218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TatD DNase domain-containing deoxyribonuclease 2 (EC 3.1.21.-). | |||||
|
ANKS1_HUMAN
|
||||||
| NC score | 0.053716 (rank : 48) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92625, Q5JYI9, Q5SYR2, Q86WQ7 | Gene names | ANKS1A, ANKS1, KIAA0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A (Odin). | |||||
|
TRPA1_MOUSE
|
||||||
| NC score | 0.050383 (rank : 49) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BLA8 | Gene names | Trpa1, Anktm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1). | |||||
|
RN5A_MOUSE
|
||||||
| NC score | 0.045907 (rank : 50) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q05921, Q9ERU7 | Gene names | Rnasel, Rns4 | |||
|
Domain Architecture |

|
|||||
| Description | 2-5A-dependent ribonuclease (EC 3.1.26.-) (2-5A-dependent RNase) (Ribonuclease L) (RNase L) (Ribonuclease 4). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.045438 (rank : 51) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
RN126_HUMAN
|
||||||
| NC score | 0.042255 (rank : 52) | θ value | 0.0252991 (rank : 33) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BV68, Q9NWX1 | Gene names | RNF126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 126. | |||||
|
RN126_MOUSE
|
||||||
| NC score | 0.041325 (rank : 53) | θ value | 0.0252991 (rank : 34) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YL2 | Gene names | Rnf126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 126. | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.036418 (rank : 54) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.029904 (rank : 55) | θ value | 0.365318 (rank : 41) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
ANR25_MOUSE
|
||||||
| NC score | 0.029729 (rank : 56) | θ value | 0.279714 (rank : 38) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BX02, Q8BLD5, Q91Z35 | Gene names | Ankrd25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25. | |||||
|
ASAH3_MOUSE
|
||||||
| NC score | 0.027967 (rank : 57) | θ value | 0.279714 (rank : 39) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R4X1 | Gene names | Asah3, Acer1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkaline ceramidase 1 (EC 3.5.1.23) (Alkaline CDase-1) (AlkCDase 1) (maCER1) (Acylsphingosine deacylase 3) (N-acylsphingosine amidohydrolase 3). | |||||
|
SC11A_HUMAN
|
||||||
| NC score | 0.024979 (rank : 58) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.024140 (rank : 59) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.021812 (rank : 60) | θ value | 0.365318 (rank : 42) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
NPAS4_MOUSE
|
||||||
| NC score | 0.020929 (rank : 61) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
NPAS4_HUMAN
|
||||||
| NC score | 0.020175 (rank : 62) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
K0562_MOUSE
|
||||||
| NC score | 0.020036 (rank : 63) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80V31, Q6ZQ95 | Gene names | Kiaa0562 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0562. | |||||
|
NELF_MOUSE
|
||||||
| NC score | 0.018905 (rank : 64) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99NF2, Q8BPT0, Q8C9R5, Q9DBF4, Q9ERZ1 | Gene names | Nelf, Jac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nasal embryonic luteinizing hormone-releasing hormone factor (Nasal embryonic LHRH factor) (Juxtasynaptic attractor of caldendrin on dendritic boutons protein) (Jacob protein). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.017093 (rank : 65) | θ value | 0.125558 (rank : 36) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
ENAM_HUMAN
|
||||||
| NC score | 0.017083 (rank : 66) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
PSD10_HUMAN
|
||||||
| NC score | 0.014041 (rank : 67) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75832 | Gene names | PSMD10 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.013295 (rank : 68) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
HRSL5_MOUSE
|
||||||
| NC score | 0.010830 (rank : 69) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CPX5, Q9CVQ2, Q9CVS0 | Gene names | Hrasls5, Hrlp5 | |||
|
Domain Architecture |

|
|||||
| Description | HRAS-like suppressor 5 (H-rev107-like protein 5). | |||||
|
CYLD_MOUSE
|
||||||
| NC score | 0.010279 (rank : 70) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80TQ2, Q80VB3, Q8BXZ3, Q8BYL9, Q8CGB0 | Gene names | Cyld, Cyld1, Kiaa0849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase CYLD (EC 3.1.2.15) (Ubiquitin thioesterase CYLD) (Ubiquitin-specific-processing protease CYLD) (Deubiquitinating enzyme CYLD). | |||||
|
C2TA_HUMAN
|
||||||
| NC score | 0.010259 (rank : 71) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
CN133_MOUSE
|
||||||
| NC score | 0.009987 (rank : 72) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BGQ1, Q8BL55, Q8CIK4, Q91YI3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf133 homolog. | |||||
|
ALEX_HUMAN
|
||||||
| NC score | 0.009634 (rank : 73) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
FBX21_MOUSE
|
||||||
| NC score | 0.009569 (rank : 74) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VDH1, Q8BHA5, Q8CHC6 | Gene names | Fbxo21, Fbx21, Kiaa0875 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 21. | |||||
|
SETX_HUMAN
|
||||||
| NC score | 0.009450 (rank : 75) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
GA2L2_MOUSE
|
||||||
| NC score | 0.009039 (rank : 76) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
REEP2_MOUSE
|
||||||
| NC score | 0.008298 (rank : 77) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCD6 | Gene names | Reep2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor expression-enhancing protein 2. | |||||
|
RBBP8_HUMAN
|
||||||
| NC score | 0.008110 (rank : 78) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99708, O75371 | Gene names | RBBP8, CTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 8 (RBBP-8) (CtBP-interacting protein) (CtIP) (Retinoblastoma-interacting protein and myosin-like) (RIM). | |||||
|
MA1A1_MOUSE
|
||||||
| NC score | 0.007322 (rank : 79) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P45700 | Gene names | Man1a1, Man1a | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IA) (Alpha-1,2-mannosidase IA) (Mannosidase alpha class 1A member 1) (Man(9)-alpha-mannosidase) (Man9-mannosidase). | |||||
|
CAN2_MOUSE
|
||||||
| NC score | 0.007308 (rank : 80) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08529, O35518, O54843 | Gene names | Capn2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (80 kDa M-calpain subunit) (CALP80). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.006780 (rank : 81) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
DGAT1_MOUSE
|
||||||
| NC score | 0.006750 (rank : 82) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2A7, Q9D7Q5 | Gene names | Dgat1, Dgat | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol O-acyltransferase 1 (EC 2.3.1.20) (Diglyceride acyltransferase). | |||||
|
CAN2_HUMAN
|
||||||
| NC score | 0.006606 (rank : 83) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17655, Q16738, Q6PJT3, Q8WU26, Q9HBB1 | Gene names | CAPN2, CANPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (Calpain large polypeptide L2). | |||||
|
UBE3C_HUMAN
|
||||||
| NC score | 0.005751 (rank : 84) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
MFN2_HUMAN
|
||||||
| NC score | 0.005279 (rank : 85) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95140, O95572, Q5JXC3, Q5JXC4, Q9H131, Q9NSX8 | Gene names | MFN2, CPRP1, KIAA0214 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane GTPase MFN2 (EC 3.6.5.-) (Mitofusin-2). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.005230 (rank : 86) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
LSD1_HUMAN
|
||||||
| NC score | 0.005085 (rank : 87) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
RIMS1_HUMAN
|
||||||
| NC score | 0.004426 (rank : 88) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
PAX6_HUMAN
|
||||||
| NC score | 0.004222 (rank : 89) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26367, Q6N006, Q99413 | Gene names | PAX6, AN2 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-6 (Oculorhombin) (Aniridia type II protein). | |||||
|
PAX6_MOUSE
|
||||||
| NC score | 0.004222 (rank : 90) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P63015, P32117, P70601, Q62222, Q64037, Q8CEI5, Q8VDB5, Q921Q8 | Gene names | Pax6, Pax-6, Sey | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paired box protein Pax-6 (Oculorhombin). | |||||
|
CABP4_HUMAN
|
||||||
| NC score | 0.003272 (rank : 91) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
TTF2_MOUSE
|
||||||
| NC score | 0.002673 (rank : 92) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
LRC47_MOUSE
|
||||||
| NC score | 0.002373 (rank : 93) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q505F5, Q3U380, Q6ZPW2, Q9CUZ0 | Gene names | Lrrc47, Kiaa1185 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 47. | |||||
|
KSR1_MOUSE
|
||||||
| NC score | 0.000152 (rank : 94) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61097, Q61648, Q78DX8 | Gene names | Ksr1, Ksr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras) (mKSR1) (Hb protein). | |||||
|
ROBO3_HUMAN
|
||||||
| NC score | -0.001079 (rank : 95) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96MS0 | Gene names | ROBO3 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Roundabout-like protein 3). | |||||
|
ZFP95_HUMAN
|
||||||
| NC score | -0.002212 (rank : 96) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2L8 | Gene names | ZFP95, KIAA1015 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 95 homolog (Zfp-95). | |||||
|
ZN672_MOUSE
|
||||||
| NC score | -0.003277 (rank : 97) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99LH4, Q8BIR7 | Gene names | Znf672, Zfp672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 672. | |||||