Please be patient as the page loads
|
TRPM8_HUMAN
|
||||||
| SwissProt Accessions | Q7Z2W7, Q6QNH9, Q8TAC3, Q8TDX8, Q9BVK1 | Gene names | TRPM8, LTRPC6, TRPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8 (Transient receptor potential-p8) (Trp-p8) (Long transient receptor potential channel 6) (LTrpC6). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TRPM2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.938026 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O94759, Q5KTC2, Q6J3P5, Q96KN6, Q96Q93 | Gene names | TRPM2, EREG1, KNP3, LTRPC2, TRPC7 | |||
|
Domain Architecture |

|
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7) (Estrogen- responsive element-associated gene 1 protein). | |||||
|
TRPM2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.935989 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q91YD4 | Gene names | Trpm2, Ltrpc2, Trpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7). | |||||
|
TRPM8_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q7Z2W7, Q6QNH9, Q8TAC3, Q8TDX8, Q9BVK1 | Gene names | TRPM8, LTRPC6, TRPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8 (Transient receptor potential-p8) (Trp-p8) (Long transient receptor potential channel 6) (LTrpC6). | |||||
|
TRPM8_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.995674 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8R4D5 | Gene names | Trpm8, Trpp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8. | |||||
|
TRPM6_MOUSE
|
||||||
| θ value | 1.3939e-101 (rank : 5) | NC score | 0.848768 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CIR4 | Gene names | Trpm6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM3_HUMAN
|
||||||
| θ value | 9.07703e-85 (rank : 6) | NC score | 0.855440 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9HCF6, Q5VW02, Q5VW03, Q5VW04, Q5W5T7, Q86SH0, Q86SH6, Q86UL0, Q86WK1, Q86WK2, Q86WK3, Q86WK4, Q86YZ9, Q86Z00, Q86Z01 | Gene names | TRPM3, KIAA1616, LTRPC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 3 (Long transient receptor potential channel 3) (LTrpC3) (Melastatin-2) (MLSN2). | |||||
|
TRPM7_MOUSE
|
||||||
| θ value | 1.50663e-63 (rank : 7) | NC score | 0.837651 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
TRPM7_HUMAN
|
||||||
| θ value | 3.35642e-63 (rank : 8) | NC score | 0.845135 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
TRPM6_HUMAN
|
||||||
| θ value | 6.77864e-56 (rank : 9) | NC score | 0.841592 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BX84, Q6VPR8, Q6VPR9, Q6VPS0, Q6VPS1, Q6VPS2 | Gene names | TRPM6, CHAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPC4_MOUSE
|
||||||
| θ value | 2.59989e-23 (rank : 10) | NC score | 0.589017 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9QUQ5, Q62350, Q9QUQ9, Q9QZC0 | Gene names | Trpc4, Trrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (Receptor- activated cation channel TRP4) (Capacitative calcium entry channel Trp4). | |||||
|
TRPC4_HUMAN
|
||||||
| θ value | 3.39556e-23 (rank : 11) | NC score | 0.590135 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
|
TRPC5_HUMAN
|
||||||
| θ value | 1.42661e-21 (rank : 12) | NC score | 0.576242 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UL62, O75233, Q5JXY8, Q9Y514 | Gene names | TRPC5, TRP5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Htrp-5) (Htrp5). | |||||
|
TRPC5_MOUSE
|
||||||
| θ value | 1.42661e-21 (rank : 13) | NC score | 0.576353 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QX29, Q61059, Q9QWT1, Q9R0D4 | Gene names | Trpc5, Trp5, Trrp5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Transient receptor protein 5) (Mtrp5) (trp-related protein 5) (Capacitative calcium entry channel 2) (CCE2). | |||||
|
TRPC2_MOUSE
|
||||||
| θ value | 4.16044e-13 (rank : 14) | NC score | 0.526969 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9R244, Q9ES59, Q9ES60, Q9R243 | Gene names | Trpc2, Trp2, Trrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 2 (TrpC2) (mTrp2). | |||||
|
TRPC3_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 15) | NC score | 0.492349 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13507, O00593, Q15660 | Gene names | TRPC3, TRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Htrp-3) (Htrp3). | |||||
|
TRPC3_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 16) | NC score | 0.497162 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QZC1, Q61058 | Gene names | Trpc3, Trp3, Trrp3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Transient receptor protein 3) (Mtrp3) (Receptor-activated cation channel TRP3) (trp-related protein 3). | |||||
|
TRPC1_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 17) | NC score | 0.471753 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P48995 | Gene names | TRPC1, TRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (TRP-1 protein). | |||||
|
TRPC1_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 18) | NC score | 0.472152 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61056, O35722 | Gene names | Trpc1, Trp1, Trrp1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (Transient receptor protein 1) (Mtrp1) (Trp-related protein 1). | |||||
|
TRPC6_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 19) | NC score | 0.459594 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
TRPC6_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 20) | NC score | 0.456381 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y210, Q9HCW3, Q9NQA8, Q9NQA9 | Gene names | TRPC6, TRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6). | |||||
|
TRPC7_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 21) | NC score | 0.452012 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WVC5 | Gene names | Trpc7, Trp7, Trrp8 | |||
|
Domain Architecture |
|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (mTRP7). | |||||
|
TRPC7_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 22) | NC score | 0.449883 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HCX4 | Gene names | TRPC7, TRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (TRP7 protein). | |||||
|
TRPV3_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 23) | NC score | 0.313768 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
TRPV3_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 24) | NC score | 0.307174 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
TRPV4_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 25) | NC score | 0.247590 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPV4_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 26) | NC score | 0.228165 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9EPK8, Q91XR5, Q9EQZ4, Q9ERZ7, Q9ES76 | Gene names | Trpv4, Trp12, Vrl2, Vroac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (Vanilloid receptor- related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPV1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 27) | NC score | 0.250432 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8NER1, Q9H0G9, Q9H303, Q9H304, Q9NQ74, Q9NY22 | Gene names | TRPV1, VR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1) (Capsaicin receptor). | |||||
|
TRPV1_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 28) | NC score | 0.244985 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q704Y3, Q5SSE1, Q5SSE2, Q5SSE4, Q5WPV5, Q68SW0 | Gene names | Trpv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1). | |||||
|
TRPV2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 29) | NC score | 0.176126 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y5S1, Q9Y670 | Gene names | TRPV2, VRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Vanilloid receptor-like protein 1) (VRL-1). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 30) | NC score | 0.064623 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
TRPV2_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 31) | NC score | 0.190889 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WTR1, Q99K71 | Gene names | Trpv2, Grc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Growth factor-regulated calcium channel) (GRC). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 32) | NC score | 0.065838 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
TRPV5_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 33) | NC score | 0.182610 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
TRPV6_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 34) | NC score | 0.192828 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91WD2 | Gene names | Trpv6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1). | |||||
|
TRPV6_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 35) | NC score | 0.209241 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9H1D0, Q8TDL3, Q8WXR8, Q96LC5, Q9H1D1, Q9H296 | Gene names | TRPV6, ECAC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1) (CaT-like) (CaT-L). | |||||
|
CAC1E_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 36) | NC score | 0.056794 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
EI24_HUMAN
|
||||||
| θ value | 0.125558 (rank : 37) | NC score | 0.082790 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14681 | Gene names | EI24, PIG8 | |||
|
Domain Architecture |
|
|||||
| Description | Etoposide-induced protein 2.4 (p53-induced protein 8). | |||||
|
EI24_MOUSE
|
||||||
| θ value | 0.125558 (rank : 38) | NC score | 0.083094 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61070 | Gene names | Ei24, Pig8 | |||
|
Domain Architecture |
|
|||||
| Description | Etoposide-induced protein 2.4 (p53-induced protein 8). | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 0.21417 (rank : 39) | NC score | 0.055364 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.049789 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
PK2L1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.124195 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
SEPP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.028672 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49908, Q6PD59, Q6PI43, Q6PI87, Q6PJF9 | Gene names | SEPP1, SELP | |||
|
Domain Architecture |

|
|||||
| Description | Selenoprotein P precursor (SeP). | |||||
|
TRPV5_MOUSE
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.180334 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P69744 | Gene names | Trpv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
CAC1A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.046936 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.003129 (rank : 72) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
NPAL2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.021339 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H841 | Gene names | NPAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NIPA-like protein 2. | |||||
|
ATP5H_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.029594 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75947, Q9H3J4 | Gene names | ATP5H | |||
|
Domain Architecture |
|
|||||
| Description | ATP synthase D chain, mitochondrial (EC 3.6.3.14). | |||||
|
MRP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.003911 (rank : 71) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92887, Q14022, Q92500, Q92798, Q99663, Q9UMS2 | Gene names | ABCC2, CMOAT, CMOAT1, CMRP, MRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Canalicular multispecific organic anion transporter 1 (ATP-binding cassette sub-family C member 2) (Multidrug resistance-associated protein 2) (Canalicular multidrug resistance protein). | |||||
|
POPD3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.013204 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ES81 | Gene names | Popdc3, Pop3 | |||
|
Domain Architecture |

|
|||||
| Description | Popeye domain-containing protein 3 (Popeye protein 3). | |||||
|
SC10A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.042117 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.038558 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
TRPA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.036854 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75762 | Gene names | TRPA1, ANKTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1) (Transformation sensitive-protein p120). | |||||
|
KCMA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.013432 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12791, Q12886, Q12917, Q12921, Q12960, Q13150, Q96LG8, Q9UBB0, Q9UQK6 | Gene names | KCNMA1, KCNMA | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (hSlo). | |||||
|
KCMA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.013436 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08460, Q64703, Q8VHF1, Q9R196 | Gene names | Kcnma1, Kcnma | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (mSlo) (mSlo1). | |||||
|
YTDC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.007596 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H6S0 | Gene names | YTHDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain-containing protein 2. | |||||
|
MPZL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.014632 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95297, Q5R332, Q8IX11, Q9BWZ3, Q9NYK4, Q9UL20 | Gene names | MPZL1, PZR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin protein zero-like protein 1 precursor (Protein zero-related). | |||||
|
SCN7A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.035930 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
DYH11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.007935 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96DT5, Q9UJ82 | Gene names | DNAH11 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 11 (Axonemal beta dynein heavy chain 11). | |||||
|
GNL3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.007196 (rank : 68) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BVP2, Q96SV6, Q96SV7, Q9UJY0 | Gene names | GNL3, E2IG3, NS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin) (E2-induced gene 3-protein) (Novel nucleolar protein 47) (NNP47). | |||||
|
GSTP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.004775 (rank : 70) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P46425, Q505C3 | Gene names | Gstp2, Gstpia | |||
|
Domain Architecture |
|
|||||
| Description | Glutathione S-transferase P 2 (EC 2.5.1.18) (GST YF-YF) (GST-piA) (GST class-pi) (Gst P2). | |||||
|
MAL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.008068 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969L2, Q6ZMD9 | Gene names | MAL2 | |||
|
Domain Architecture |
|
|||||
| Description | Protein MAL2. | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | -0.000655 (rank : 73) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
PKD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.077792 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
SC10A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.038364 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
TRXR2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.005169 (rank : 69) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JLT4, Q91YX4, Q9JHA7, Q9JMH5 | Gene names | Txnrd2, Trxr2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 2, mitochondrial precursor (EC 1.8.1.9) (TR3). | |||||
|
EF2K_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.128503 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00418 | Gene names | EEF2K | |||
|
Domain Architecture |
|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
EF2K_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.133589 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08796 | Gene names | Eef2k | |||
|
Domain Architecture |
|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
NUD9P_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.157307 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N2Z3 | Gene names | NUDT9P1, C10orf98 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative nucleoside diphosphate-linked moiety X motif 9P1. | |||||
|
NUDT9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.207825 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BW91, Q8NBN1, Q8NCB9, Q8NG25 | Gene names | NUDT9, NUDT10 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate- linked moiety X motif 9) (Nudix motif 9). | |||||
|
NUDT9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.209190 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVU5, Q3TZ68, Q8K1J4 | Gene names | Nudt9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate- linked moiety X motif 9) (Nudix motif 9). | |||||
|
PK2L2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.078610 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.086985 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PKD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.068638 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
TRPM8_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q7Z2W7, Q6QNH9, Q8TAC3, Q8TDX8, Q9BVK1 | Gene names | TRPM8, LTRPC6, TRPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8 (Transient receptor potential-p8) (Trp-p8) (Long transient receptor potential channel 6) (LTrpC6). | |||||
|
TRPM8_MOUSE
|
||||||
| NC score | 0.995674 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8R4D5 | Gene names | Trpm8, Trpp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8. | |||||
|
TRPM2_HUMAN
|
||||||
| NC score | 0.938026 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O94759, Q5KTC2, Q6J3P5, Q96KN6, Q96Q93 | Gene names | TRPM2, EREG1, KNP3, LTRPC2, TRPC7 | |||
|
Domain Architecture |

|
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7) (Estrogen- responsive element-associated gene 1 protein). | |||||
|
TRPM2_MOUSE
|
||||||
| NC score | 0.935989 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q91YD4 | Gene names | Trpm2, Ltrpc2, Trpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7). | |||||
|
TRPM3_HUMAN
|
||||||
| NC score | 0.855440 (rank : 5) | θ value | 9.07703e-85 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9HCF6, Q5VW02, Q5VW03, Q5VW04, Q5W5T7, Q86SH0, Q86SH6, Q86UL0, Q86WK1, Q86WK2, Q86WK3, Q86WK4, Q86YZ9, Q86Z00, Q86Z01 | Gene names | TRPM3, KIAA1616, LTRPC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 3 (Long transient receptor potential channel 3) (LTrpC3) (Melastatin-2) (MLSN2). | |||||
|
TRPM6_MOUSE
|
||||||
| NC score | 0.848768 (rank : 6) | θ value | 1.3939e-101 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CIR4 | Gene names | Trpm6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM7_HUMAN
|
||||||
| NC score | 0.845135 (rank : 7) | θ value | 3.35642e-63 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
TRPM6_HUMAN
|
||||||
| NC score | 0.841592 (rank : 8) | θ value | 6.77864e-56 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BX84, Q6VPR8, Q6VPR9, Q6VPS0, Q6VPS1, Q6VPS2 | Gene names | TRPM6, CHAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM7_MOUSE
|
||||||
| NC score | 0.837651 (rank : 9) | θ value | 1.50663e-63 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
TRPC4_HUMAN
|
||||||
| NC score | 0.590135 (rank : 10) | θ value | 3.39556e-23 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
|
TRPC4_MOUSE
|
||||||
| NC score | 0.589017 (rank : 11) | θ value | 2.59989e-23 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9QUQ5, Q62350, Q9QUQ9, Q9QZC0 | Gene names | Trpc4, Trrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (Receptor- activated cation channel TRP4) (Capacitative calcium entry channel Trp4). | |||||
|
TRPC5_MOUSE
|
||||||
| NC score | 0.576353 (rank : 12) | θ value | 1.42661e-21 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QX29, Q61059, Q9QWT1, Q9R0D4 | Gene names | Trpc5, Trp5, Trrp5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Transient receptor protein 5) (Mtrp5) (trp-related protein 5) (Capacitative calcium entry channel 2) (CCE2). | |||||
|
TRPC5_HUMAN
|
||||||
| NC score | 0.576242 (rank : 13) | θ value | 1.42661e-21 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UL62, O75233, Q5JXY8, Q9Y514 | Gene names | TRPC5, TRP5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Htrp-5) (Htrp5). | |||||
|
TRPC2_MOUSE
|
||||||
| NC score | 0.526969 (rank : 14) | θ value | 4.16044e-13 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9R244, Q9ES59, Q9ES60, Q9R243 | Gene names | Trpc2, Trp2, Trrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 2 (TrpC2) (mTrp2). | |||||
|
TRPC3_MOUSE
|
||||||
| NC score | 0.497162 (rank : 15) | θ value | 3.29651e-10 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QZC1, Q61058 | Gene names | Trpc3, Trp3, Trrp3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Transient receptor protein 3) (Mtrp3) (Receptor-activated cation channel TRP3) (trp-related protein 3). | |||||
|
TRPC3_HUMAN
|
||||||
| NC score | 0.492349 (rank : 16) | θ value | 1.9326e-10 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13507, O00593, Q15660 | Gene names | TRPC3, TRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Htrp-3) (Htrp3). | |||||
|
TRPC1_MOUSE
|
||||||
| NC score | 0.472152 (rank : 17) | θ value | 2.79066e-09 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61056, O35722 | Gene names | Trpc1, Trp1, Trrp1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (Transient receptor protein 1) (Mtrp1) (Trp-related protein 1). | |||||
|
TRPC1_HUMAN
|
||||||
| NC score | 0.471753 (rank : 18) | θ value | 2.79066e-09 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P48995 | Gene names | TRPC1, TRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (TRP-1 protein). | |||||
|
TRPC6_MOUSE
|
||||||
| NC score | 0.459594 (rank : 19) | θ value | 1.38499e-08 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
TRPC6_HUMAN
|
||||||
| NC score | 0.456381 (rank : 20) | θ value | 4.0297e-08 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y210, Q9HCW3, Q9NQA8, Q9NQA9 | Gene names | TRPC6, TRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6). | |||||
|
TRPC7_MOUSE
|
||||||
| NC score | 0.452012 (rank : 21) | θ value | 4.0297e-08 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WVC5 | Gene names | Trpc7, Trp7, Trrp8 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (mTRP7). | |||||
|
TRPC7_HUMAN
|
||||||
| NC score | 0.449883 (rank : 22) | θ value | 1.99992e-07 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HCX4 | Gene names | TRPC7, TRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (TRP7 protein). | |||||
|
TRPV3_HUMAN
|
||||||
| NC score | 0.313768 (rank : 23) | θ value | 2.21117e-06 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
TRPV3_MOUSE
|
||||||
| NC score | 0.307174 (rank : 24) | θ value | 1.43324e-05 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
TRPV1_HUMAN
|
||||||
| NC score | 0.250432 (rank : 25) | θ value | 0.00228821 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8NER1, Q9H0G9, Q9H303, Q9H304, Q9NQ74, Q9NY22 | Gene names | TRPV1, VR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1) (Capsaicin receptor). | |||||
|
TRPV4_HUMAN
|
||||||
| NC score | 0.247590 (rank : 26) | θ value | 0.000158464 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPV1_MOUSE
|
||||||
| NC score | 0.244985 (rank : 27) | θ value | 0.00228821 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q704Y3, Q5SSE1, Q5SSE2, Q5SSE4, Q5WPV5, Q68SW0 | Gene names | Trpv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1). | |||||
|
TRPV4_MOUSE
|
||||||
| NC score | 0.228165 (rank : 28) | θ value | 0.00134147 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9EPK8, Q91XR5, Q9EQZ4, Q9ERZ7, Q9ES76 | Gene names | Trpv4, Trp12, Vrl2, Vroac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (Vanilloid receptor- related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPV6_HUMAN
|
||||||
| NC score | 0.209241 (rank : 29) | θ value | 0.0330416 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9H1D0, Q8TDL3, Q8WXR8, Q96LC5, Q9H1D1, Q9H296 | Gene names | TRPV6, ECAC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1) (CaT-like) (CaT-L). | |||||
|
NUDT9_MOUSE
|
||||||
| NC score | 0.209190 (rank : 30) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVU5, Q3TZ68, Q8K1J4 | Gene names | Nudt9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate- linked moiety X motif 9) (Nudix motif 9). | |||||
|
NUDT9_HUMAN
|
||||||
| NC score | 0.207825 (rank : 31) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BW91, Q8NBN1, Q8NCB9, Q8NG25 | Gene names | NUDT9, NUDT10 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate- linked moiety X motif 9) (Nudix motif 9). | |||||
|
TRPV6_MOUSE
|
||||||
| NC score | 0.192828 (rank : 32) | θ value | 0.0252991 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91WD2 | Gene names | Trpv6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1). | |||||
|
TRPV2_MOUSE
|
||||||
| NC score | 0.190889 (rank : 33) | θ value | 0.0148317 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WTR1, Q99K71 | Gene names | Trpv2, Grc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Growth factor-regulated calcium channel) (GRC). | |||||
|
TRPV5_HUMAN
|
||||||
| NC score | 0.182610 (rank : 34) | θ value | 0.0252991 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
TRPV5_MOUSE
|
||||||
| NC score | 0.180334 (rank : 35) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P69744 | Gene names | Trpv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
TRPV2_HUMAN
|
||||||
| NC score | 0.176126 (rank : 36) | θ value | 0.0113563 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y5S1, Q9Y670 | Gene names | TRPV2, VRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Vanilloid receptor-like protein 1) (VRL-1). | |||||
|
NUD9P_HUMAN
|
||||||
| NC score | 0.157307 (rank : 37) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N2Z3 | Gene names | NUDT9P1, C10orf98 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative nucleoside diphosphate-linked moiety X motif 9P1. | |||||
|
EF2K_MOUSE
|
||||||
| NC score | 0.133589 (rank : 38) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08796 | Gene names | Eef2k | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
EF2K_HUMAN
|
||||||
| NC score | 0.128503 (rank : 39) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00418 | Gene names | EEF2K | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
PK2L1_HUMAN
|
||||||
| NC score | 0.124195 (rank : 40) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PK2L2_MOUSE
|
||||||
| NC score | 0.086985 (rank : 41) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
EI24_MOUSE
|
||||||
| NC score | 0.083094 (rank : 42) | θ value | 0.125558 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61070 | Gene names | Ei24, Pig8 | |||
|
Domain Architecture |

|
|||||
| Description | Etoposide-induced protein 2.4 (p53-induced protein 8). | |||||
|
EI24_HUMAN
|
||||||
| NC score | 0.082790 (rank : 43) | θ value | 0.125558 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14681 | Gene names | EI24, PIG8 | |||
|
Domain Architecture |

|
|||||
| Description | Etoposide-induced protein 2.4 (p53-induced protein 8). | |||||
|
PK2L2_HUMAN
|
||||||
| NC score | 0.078610 (rank : 44) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PKD2_HUMAN
|
||||||
| NC score | 0.077792 (rank : 45) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
PKD2_MOUSE
|
||||||
| NC score | 0.068638 (rank : 46) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.065838 (rank : 47) | θ value | 0.0193708 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.064623 (rank : 48) | θ value | 0.0148317 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1E_HUMAN
|
||||||
| NC score | 0.056794 (rank : 49) | θ value | 0.0961366 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.055364 (rank : 50) | θ value | 0.21417 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.049789 (rank : 51) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1A_HUMAN
|
||||||
| NC score | 0.046936 (rank : 52) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
SC10A_MOUSE
|
||||||
| NC score | 0.042117 (rank : 53) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.038558 (rank : 54) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SC10A_HUMAN
|
||||||
| NC score | 0.038364 (rank : 55) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
TRPA1_HUMAN
|
||||||
| NC score | 0.036854 (rank : 56) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75762 | Gene names | TRPA1, ANKTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1) (Transformation sensitive-protein p120). | |||||
|
SCN7A_HUMAN
|
||||||
| NC score | 0.035930 (rank : 57) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
ATP5H_HUMAN
|
||||||
| NC score | 0.029594 (rank : 58) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75947, Q9H3J4 | Gene names | ATP5H | |||
|
Domain Architecture |

|
|||||
| Description | ATP synthase D chain, mitochondrial (EC 3.6.3.14). | |||||
|
SEPP1_HUMAN
|
||||||
| NC score | 0.028672 (rank : 59) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49908, Q6PD59, Q6PI43, Q6PI87, Q6PJF9 | Gene names | SEPP1, SELP | |||
|
Domain Architecture |

|
|||||
| Description | Selenoprotein P precursor (SeP). | |||||
|
NPAL2_HUMAN
|
||||||
| NC score | 0.021339 (rank : 60) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H841 | Gene names | NPAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NIPA-like protein 2. | |||||
|
MPZL1_HUMAN
|
||||||
| NC score | 0.014632 (rank : 61) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95297, Q5R332, Q8IX11, Q9BWZ3, Q9NYK4, Q9UL20 | Gene names | MPZL1, PZR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin protein zero-like protein 1 precursor (Protein zero-related). | |||||
|
KCMA1_MOUSE
|
||||||
| NC score | 0.013436 (rank : 62) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08460, Q64703, Q8VHF1, Q9R196 | Gene names | Kcnma1, Kcnma | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (mSlo) (mSlo1). | |||||
|
KCMA1_HUMAN
|
||||||
| NC score | 0.013432 (rank : 63) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12791, Q12886, Q12917, Q12921, Q12960, Q13150, Q96LG8, Q9UBB0, Q9UQK6 | Gene names | KCNMA1, KCNMA | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (hSlo). | |||||
|
POPD3_MOUSE
|
||||||
| NC score | 0.013204 (rank : 64) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ES81 | Gene names | Popdc3, Pop3 | |||
|
Domain Architecture |

|
|||||
| Description | Popeye domain-containing protein 3 (Popeye protein 3). | |||||
|
MAL2_HUMAN
|
||||||
| NC score | 0.008068 (rank : 65) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969L2, Q6ZMD9 | Gene names | MAL2 | |||
|
Domain Architecture |
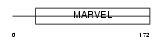
|
|||||
| Description | Protein MAL2. | |||||
|
DYH11_HUMAN
|
||||||
| NC score | 0.007935 (rank : 66) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96DT5, Q9UJ82 | Gene names | DNAH11 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 11 (Axonemal beta dynein heavy chain 11). | |||||
|
YTDC2_HUMAN
|
||||||
| NC score | 0.007596 (rank : 67) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H6S0 | Gene names | YTHDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain-containing protein 2. | |||||
|
GNL3_HUMAN
|
||||||
| NC score | 0.007196 (rank : 68) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BVP2, Q96SV6, Q96SV7, Q9UJY0 | Gene names | GNL3, E2IG3, NS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin) (E2-induced gene 3-protein) (Novel nucleolar protein 47) (NNP47). | |||||
|
TRXR2_MOUSE
|
||||||
| NC score | 0.005169 (rank : 69) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JLT4, Q91YX4, Q9JHA7, Q9JMH5 | Gene names | Txnrd2, Trxr2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 2, mitochondrial precursor (EC 1.8.1.9) (TR3). | |||||
|
GSTP2_MOUSE
|
||||||
| NC score | 0.004775 (rank : 70) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P46425, Q505C3 | Gene names | Gstp2, Gstpia | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione S-transferase P 2 (EC 2.5.1.18) (GST YF-YF) (GST-piA) (GST class-pi) (Gst P2). | |||||
|
MRP2_HUMAN
|
||||||
| NC score | 0.003911 (rank : 71) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92887, Q14022, Q92500, Q92798, Q99663, Q9UMS2 | Gene names | ABCC2, CMOAT, CMOAT1, CMRP, MRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Canalicular multispecific organic anion transporter 1 (ATP-binding cassette sub-family C member 2) (Multidrug resistance-associated protein 2) (Canalicular multidrug resistance protein). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.003129 (rank : 72) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | -0.000655 (rank : 73) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||