Please be patient as the page loads
|
TRPM6_MOUSE
|
||||||
| SwissProt Accessions | Q8CIR4 | Gene names | Trpm6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TRPM3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.959760 (rank : 5) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HCF6, Q5VW02, Q5VW03, Q5VW04, Q5W5T7, Q86SH0, Q86SH6, Q86UL0, Q86WK1, Q86WK2, Q86WK3, Q86WK4, Q86YZ9, Q86Z00, Q86Z01 | Gene names | TRPM3, KIAA1616, LTRPC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 3 (Long transient receptor potential channel 3) (LTrpC3) (Melastatin-2) (MLSN2). | |||||
|
TRPM6_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.990346 (rank : 2) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BX84, Q6VPR8, Q6VPR9, Q6VPS0, Q6VPS1, Q6VPS2 | Gene names | TRPM6, CHAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM6_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8CIR4 | Gene names | Trpm6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM7_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.977476 (rank : 4) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
TRPM7_MOUSE
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.978272 (rank : 3) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
TRPM8_HUMAN
|
||||||
| θ value | 1.3939e-101 (rank : 6) | NC score | 0.848768 (rank : 7) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7Z2W7, Q6QNH9, Q8TAC3, Q8TDX8, Q9BVK1 | Gene names | TRPM8, LTRPC6, TRPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8 (Transient receptor potential-p8) (Trp-p8) (Long transient receptor potential channel 6) (LTrpC6). | |||||
|
TRPM2_MOUSE
|
||||||
| θ value | 9.056e-93 (rank : 7) | NC score | 0.851060 (rank : 6) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91YD4 | Gene names | Trpm2, Ltrpc2, Trpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7). | |||||
|
TRPM2_HUMAN
|
||||||
| θ value | 1.44581e-90 (rank : 8) | NC score | 0.847341 (rank : 8) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O94759, Q5KTC2, Q6J3P5, Q96KN6, Q96Q93 | Gene names | TRPM2, EREG1, KNP3, LTRPC2, TRPC7 | |||
|
Domain Architecture |

|
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7) (Estrogen- responsive element-associated gene 1 protein). | |||||
|
TRPM8_MOUSE
|
||||||
| θ value | 2.18062e-54 (rank : 9) | NC score | 0.843088 (rank : 9) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R4D5 | Gene names | Trpm8, Trpp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8. | |||||
|
TRPC4_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 10) | NC score | 0.486156 (rank : 11) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QUQ5, Q62350, Q9QUQ9, Q9QZC0 | Gene names | Trpc4, Trrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (Receptor- activated cation channel TRP4) (Capacitative calcium entry channel Trp4). | |||||
|
TRPC4_HUMAN
|
||||||
| θ value | 5.25075e-16 (rank : 11) | NC score | 0.487317 (rank : 10) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
|
TRPC5_HUMAN
|
||||||
| θ value | 1.68911e-14 (rank : 12) | NC score | 0.470178 (rank : 13) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UL62, O75233, Q5JXY8, Q9Y514 | Gene names | TRPC5, TRP5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Htrp-5) (Htrp5). | |||||
|
TRPC5_MOUSE
|
||||||
| θ value | 1.68911e-14 (rank : 13) | NC score | 0.470499 (rank : 12) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QX29, Q61059, Q9QWT1, Q9R0D4 | Gene names | Trpc5, Trp5, Trrp5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Transient receptor protein 5) (Mtrp5) (trp-related protein 5) (Capacitative calcium entry channel 2) (CCE2). | |||||
|
EF2K_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 14) | NC score | 0.343178 (rank : 23) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08796 | Gene names | Eef2k | |||
|
Domain Architecture |
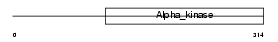
|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
EF2K_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 15) | NC score | 0.334990 (rank : 24) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00418 | Gene names | EEF2K | |||
|
Domain Architecture |
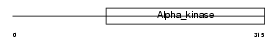
|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
TRPC2_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 16) | NC score | 0.435137 (rank : 14) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9R244, Q9ES59, Q9ES60, Q9R243 | Gene names | Trpc2, Trp2, Trrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 2 (TrpC2) (mTrp2). | |||||
|
TRPC3_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 17) | NC score | 0.405864 (rank : 15) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QZC1, Q61058 | Gene names | Trpc3, Trp3, Trrp3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Transient receptor protein 3) (Mtrp3) (Receptor-activated cation channel TRP3) (trp-related protein 3). | |||||
|
TRPC7_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 18) | NC score | 0.366325 (rank : 20) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HCX4 | Gene names | TRPC7, TRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (TRP7 protein). | |||||
|
TRPC7_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 19) | NC score | 0.367226 (rank : 19) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9WVC5 | Gene names | Trpc7, Trp7, Trrp8 | |||
|
Domain Architecture |
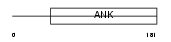
|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (mTRP7). | |||||
|
TRPC6_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 20) | NC score | 0.374651 (rank : 18) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y210, Q9HCW3, Q9NQA8, Q9NQA9 | Gene names | TRPC6, TRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6). | |||||
|
TRPC6_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 21) | NC score | 0.375315 (rank : 17) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
TRPC3_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 22) | NC score | 0.394975 (rank : 16) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13507, O00593, Q15660 | Gene names | TRPC3, TRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Htrp-3) (Htrp3). | |||||
|
TRPV3_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 23) | NC score | 0.222551 (rank : 25) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
TRPV3_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 24) | NC score | 0.221802 (rank : 26) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
TRPC1_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 25) | NC score | 0.346053 (rank : 22) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P48995 | Gene names | TRPC1, TRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (TRP-1 protein). | |||||
|
TRPC1_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 26) | NC score | 0.346213 (rank : 21) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61056, O35722 | Gene names | Trpc1, Trp1, Trrp1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (Transient receptor protein 1) (Mtrp1) (Trp-related protein 1). | |||||
|
TRPV5_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 27) | NC score | 0.142239 (rank : 30) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
TRPV6_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 28) | NC score | 0.158423 (rank : 27) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H1D0, Q8TDL3, Q8WXR8, Q96LC5, Q9H1D1, Q9H296 | Gene names | TRPV6, ECAC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1) (CaT-like) (CaT-L). | |||||
|
TRPV6_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 29) | NC score | 0.144289 (rank : 29) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91WD2 | Gene names | Trpv6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1). | |||||
|
TRPV5_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 30) | NC score | 0.135403 (rank : 34) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P69744 | Gene names | Trpv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
CNGA1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 31) | NC score | 0.020231 (rank : 47) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.008680 (rank : 69) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
UBP37_HUMAN
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.016825 (rank : 50) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
FRM4B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.016908 (rank : 49) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2L6, Q8TAI3 | Gene names | FRMD4B, KIAA1013 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.012409 (rank : 58) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
CNGA1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.023862 (rank : 46) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
COHA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.008120 (rank : 74) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UMD9, Q02802, Q99018, Q9NQK9, Q9UC14 | Gene names | COL17A1, BP180, BPAG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
LRSM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.010371 (rank : 66) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
NUCL_MOUSE
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.008629 (rank : 70) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
CBPB2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.009012 (rank : 68) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHH6, Q5EBI3, Q9QZF0 | Gene names | Cpb2, Tafi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase B2 precursor (EC 3.4.17.20) (Carboxypeptidase U) (CPU) (Thrombin-activable fibrinolysis inhibitor) (TAFI) (Carboxypeptidase R) (CPR). | |||||
|
DMD_MOUSE
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.010958 (rank : 65) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
INP4A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.013718 (rank : 54) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPW0, Q8CBJ8 | Gene names | Inpp4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type I inositol-3,4-bisphosphate 4-phosphatase (EC 3.1.3.66) (Inositol polyphosphate 4-phosphatase type I). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.032488 (rank : 43) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.015445 (rank : 52) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | -0.001540 (rank : 88) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
FANCE_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.025897 (rank : 45) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HB96, Q4ZGH2 | Gene names | FANCE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group E protein (Protein FACE). | |||||
|
FRM4B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.014005 (rank : 53) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q920B0, Q3V1S1, Q9ESP9 | Gene names | Frmd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1) (Golgi- associated band 4.1-like protein) (GOBLIN). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.012876 (rank : 57) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.005242 (rank : 84) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.012908 (rank : 56) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
TRPV1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.146177 (rank : 28) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NER1, Q9H0G9, Q9H303, Q9H304, Q9NQ74, Q9NY22 | Gene names | TRPV1, VR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1) (Capsaicin receptor). | |||||
|
ANLN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.013480 (rank : 55) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
CCD91_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.012053 (rank : 59) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 699 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D8L5, Q8R3N8, Q9D9E8 | Gene names | Ccdc91, Ggabp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.011976 (rank : 60) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
NMDE1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.005264 (rank : 83) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12879, O00669 | Gene names | GRIN2A | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A) (hNR2A). | |||||
|
RF1M_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.016038 (rank : 51) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75570, Q5T6Y5, Q8IUQ6 | Gene names | MTRF1 | |||
|
Domain Architecture |
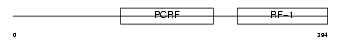
|
|||||
| Description | Peptide chain release factor 1, mitochondrial precursor (MRF-1). | |||||
|
CF170_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.017309 (rank : 48) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96NH3, Q6ZMY4, Q6ZUR7, Q8NB47 | Gene names | C6orf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf170. | |||||
|
MYPT1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.007790 (rank : 75) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
MYPT1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.011694 (rank : 62) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.027366 (rank : 44) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
ZN142_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | -0.002450 (rank : 89) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
CCD46_MOUSE
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.006956 (rank : 76) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CCD93_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.008195 (rank : 73) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7TQK5, Q3TX53 | Gene names | Ccdc93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 93. | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.005371 (rank : 82) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
MTERF_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.011270 (rank : 64) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CHZ9, Q63ZX5, Q8C7T1 | Gene names | Mterf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor, mitochondrial precursor (mTERF) (Mitochondrial transcription termination factor 1). | |||||
|
SLC2B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.011729 (rank : 61) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NEV8, Q9Y4D6 | Gene names | EXPH5, KIAA0624, SLAC2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Slp homolog lacking C2 domains b (Exophilin-5). | |||||
|
STXB4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.008342 (rank : 72) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 616 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6ZWJ1, Q8IVZ5 | Gene names | STXBP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.004044 (rank : 86) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
BICD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.005971 (rank : 80) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
CP135_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.004656 (rank : 85) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
DOS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.011379 (rank : 63) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N350, O43385 | Gene names | DOS, C19orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dos. | |||||
|
EDN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.008548 (rank : 71) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48299, Q543L0 | Gene names | Edn3 | |||
|
Domain Architecture |
|
|||||
| Description | Endothelin-3 precursor (ET-3) (Preproendothelin-3) (PPET3). | |||||
|
FA8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.003533 (rank : 87) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q06194 | Gene names | F8, Cf8, F8c | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component). | |||||
|
NCKPL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.006299 (rank : 78) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55160 | Gene names | NCKAP1L, HEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nck-associated protein 1-like (Membrane-associated protein HEM-1) (Hematopoietic protein 1). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.009014 (rank : 67) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
PF21B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.005435 (rank : 81) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PRGC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.005991 (rank : 79) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86YN6, Q86YN3, Q86YN4, Q86YN5, Q8N1N9, Q8TDE4, Q8TDE5 | Gene names | PPARGC1B, PERC, PGC1, PGC1B, PPARGC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta) (PGC-1-related estrogen receptor alpha coactivator) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
WWTR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.006862 (rank : 77) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EPK5, Q99KI4 | Gene names | Wwtr1, Taz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
NUD9P_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.103483 (rank : 38) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N2Z3 | Gene names | NUDT9P1, C10orf98 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative nucleoside diphosphate-linked moiety X motif 9P1. | |||||
|
NUDT9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.136135 (rank : 33) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BW91, Q8NBN1, Q8NCB9, Q8NG25 | Gene names | NUDT9, NUDT10 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate- linked moiety X motif 9) (Nudix motif 9). | |||||
|
NUDT9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.136933 (rank : 32) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BVU5, Q3TZ68, Q8K1J4 | Gene names | Nudt9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate- linked moiety X motif 9) (Nudix motif 9). | |||||
|
PK2L1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.093806 (rank : 39) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PK2L2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.057296 (rank : 42) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.069253 (rank : 41) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
TRPV1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.139831 (rank : 31) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q704Y3, Q5SSE1, Q5SSE2, Q5SSE4, Q5WPV5, Q68SW0 | Gene names | Trpv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1). | |||||
|
TRPV2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.081510 (rank : 40) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5S1, Q9Y670 | Gene names | TRPV2, VRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Vanilloid receptor-like protein 1) (VRL-1). | |||||
|
TRPV2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.105611 (rank : 37) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WTR1, Q99K71 | Gene names | Trpv2, Grc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Growth factor-regulated calcium channel) (GRC). | |||||
|
TRPV4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.133036 (rank : 35) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPV4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.119096 (rank : 36) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9EPK8, Q91XR5, Q9EQZ4, Q9ERZ7, Q9ES76 | Gene names | Trpv4, Trp12, Vrl2, Vroac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (Vanilloid receptor- related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPM6_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8CIR4 | Gene names | Trpm6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM6_HUMAN
|
||||||
| NC score | 0.990346 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BX84, Q6VPR8, Q6VPR9, Q6VPS0, Q6VPS1, Q6VPS2 | Gene names | TRPM6, CHAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM7_MOUSE
|
||||||
| NC score | 0.978272 (rank : 3) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
TRPM7_HUMAN
|
||||||
| NC score | 0.977476 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
TRPM3_HUMAN
|
||||||
| NC score | 0.959760 (rank : 5) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HCF6, Q5VW02, Q5VW03, Q5VW04, Q5W5T7, Q86SH0, Q86SH6, Q86UL0, Q86WK1, Q86WK2, Q86WK3, Q86WK4, Q86YZ9, Q86Z00, Q86Z01 | Gene names | TRPM3, KIAA1616, LTRPC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 3 (Long transient receptor potential channel 3) (LTrpC3) (Melastatin-2) (MLSN2). | |||||
|
TRPM2_MOUSE
|
||||||
| NC score | 0.851060 (rank : 6) | θ value | 9.056e-93 (rank : 7) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91YD4 | Gene names | Trpm2, Ltrpc2, Trpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7). | |||||
|
TRPM8_HUMAN
|
||||||
| NC score | 0.848768 (rank : 7) | θ value | 1.3939e-101 (rank : 6) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7Z2W7, Q6QNH9, Q8TAC3, Q8TDX8, Q9BVK1 | Gene names | TRPM8, LTRPC6, TRPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8 (Transient receptor potential-p8) (Trp-p8) (Long transient receptor potential channel 6) (LTrpC6). | |||||
|
TRPM2_HUMAN
|
||||||
| NC score | 0.847341 (rank : 8) | θ value | 1.44581e-90 (rank : 8) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O94759, Q5KTC2, Q6J3P5, Q96KN6, Q96Q93 | Gene names | TRPM2, EREG1, KNP3, LTRPC2, TRPC7 | |||
|
Domain Architecture |

|
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7) (Estrogen- responsive element-associated gene 1 protein). | |||||
|
TRPM8_MOUSE
|
||||||
| NC score | 0.843088 (rank : 9) | θ value | 2.18062e-54 (rank : 9) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R4D5 | Gene names | Trpm8, Trpp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8. | |||||
|
TRPC4_HUMAN
|
||||||
| NC score | 0.487317 (rank : 10) | θ value | 5.25075e-16 (rank : 11) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
|
TRPC4_MOUSE
|
||||||
| NC score | 0.486156 (rank : 11) | θ value | 4.02038e-16 (rank : 10) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QUQ5, Q62350, Q9QUQ9, Q9QZC0 | Gene names | Trpc4, Trrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (Receptor- activated cation channel TRP4) (Capacitative calcium entry channel Trp4). | |||||
|
TRPC5_MOUSE
|
||||||
| NC score | 0.470499 (rank : 12) | θ value | 1.68911e-14 (rank : 13) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QX29, Q61059, Q9QWT1, Q9R0D4 | Gene names | Trpc5, Trp5, Trrp5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Transient receptor protein 5) (Mtrp5) (trp-related protein 5) (Capacitative calcium entry channel 2) (CCE2). | |||||
|
TRPC5_HUMAN
|
||||||
| NC score | 0.470178 (rank : 13) | θ value | 1.68911e-14 (rank : 12) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UL62, O75233, Q5JXY8, Q9Y514 | Gene names | TRPC5, TRP5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Htrp-5) (Htrp5). | |||||
|
TRPC2_MOUSE
|
||||||
| NC score | 0.435137 (rank : 14) | θ value | 2.36244e-08 (rank : 16) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9R244, Q9ES59, Q9ES60, Q9R243 | Gene names | Trpc2, Trp2, Trrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 2 (TrpC2) (mTrp2). | |||||
|
TRPC3_MOUSE
|
||||||
| NC score | 0.405864 (rank : 15) | θ value | 2.36244e-08 (rank : 17) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QZC1, Q61058 | Gene names | Trpc3, Trp3, Trrp3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Transient receptor protein 3) (Mtrp3) (Receptor-activated cation channel TRP3) (trp-related protein 3). | |||||
|
TRPC3_HUMAN
|
||||||
| NC score | 0.394975 (rank : 16) | θ value | 5.81887e-07 (rank : 22) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13507, O00593, Q15660 | Gene names | TRPC3, TRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Htrp-3) (Htrp3). | |||||
|
TRPC6_MOUSE
|
||||||
| NC score | 0.375315 (rank : 17) | θ value | 2.61198e-07 (rank : 21) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
TRPC6_HUMAN
|
||||||
| NC score | 0.374651 (rank : 18) | θ value | 2.61198e-07 (rank : 20) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y210, Q9HCW3, Q9NQA8, Q9NQA9 | Gene names | TRPC6, TRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6). | |||||
|
TRPC7_MOUSE
|
||||||
| NC score | 0.367226 (rank : 19) | θ value | 1.99992e-07 (rank : 19) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9WVC5 | Gene names | Trpc7, Trp7, Trrp8 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (mTRP7). | |||||
|
TRPC7_HUMAN
|
||||||
| NC score | 0.366325 (rank : 20) | θ value | 1.99992e-07 (rank : 18) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HCX4 | Gene names | TRPC7, TRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (TRP7 protein). | |||||
|
TRPC1_MOUSE
|
||||||
| NC score | 0.346213 (rank : 21) | θ value | 0.000461057 (rank : 26) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61056, O35722 | Gene names | Trpc1, Trp1, Trrp1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (Transient receptor protein 1) (Mtrp1) (Trp-related protein 1). | |||||
|
TRPC1_HUMAN
|
||||||
| NC score | 0.346053 (rank : 22) | θ value | 0.000461057 (rank : 25) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P48995 | Gene names | TRPC1, TRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (TRP-1 protein). | |||||
|
EF2K_MOUSE
|
||||||
| NC score | 0.343178 (rank : 23) | θ value | 4.59992e-12 (rank : 14) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08796 | Gene names | Eef2k | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
EF2K_HUMAN
|
||||||
| NC score | 0.334990 (rank : 24) | θ value | 5.08577e-11 (rank : 15) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00418 | Gene names | EEF2K | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
TRPV3_MOUSE
|
||||||
| NC score | 0.222551 (rank : 25) | θ value | 3.19293e-05 (rank : 23) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
TRPV3_HUMAN
|
||||||
| NC score | 0.221802 (rank : 26) | θ value | 7.1131e-05 (rank : 24) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
TRPV6_HUMAN
|
||||||
| NC score | 0.158423 (rank : 27) | θ value | 0.0148317 (rank : 28) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H1D0, Q8TDL3, Q8WXR8, Q96LC5, Q9H1D1, Q9H296 | Gene names | TRPV6, ECAC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1) (CaT-like) (CaT-L). | |||||
|
TRPV1_HUMAN
|
||||||
| NC score | 0.146177 (rank : 28) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NER1, Q9H0G9, Q9H303, Q9H304, Q9NQ74, Q9NY22 | Gene names | TRPV1, VR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1) (Capsaicin receptor). | |||||
|
TRPV6_MOUSE
|
||||||
| NC score | 0.144289 (rank : 29) | θ value | 0.0252991 (rank : 29) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91WD2 | Gene names | Trpv6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1). | |||||
|
TRPV5_HUMAN
|
||||||
| NC score | 0.142239 (rank : 30) | θ value | 0.00869519 (rank : 27) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
TRPV1_MOUSE
|
||||||
| NC score | 0.139831 (rank : 31) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q704Y3, Q5SSE1, Q5SSE2, Q5SSE4, Q5WPV5, Q68SW0 | Gene names | Trpv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1). | |||||
|
NUDT9_MOUSE
|
||||||
| NC score | 0.136933 (rank : 32) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BVU5, Q3TZ68, Q8K1J4 | Gene names | Nudt9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate- linked moiety X motif 9) (Nudix motif 9). | |||||
|
NUDT9_HUMAN
|
||||||
| NC score | 0.136135 (rank : 33) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BW91, Q8NBN1, Q8NCB9, Q8NG25 | Gene names | NUDT9, NUDT10 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate- linked moiety X motif 9) (Nudix motif 9). | |||||
|
TRPV5_MOUSE
|
||||||
| NC score | 0.135403 (rank : 34) | θ value | 0.0563607 (rank : 30) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P69744 | Gene names | Trpv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
TRPV4_HUMAN
|
||||||
| NC score | 0.133036 (rank : 35) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPV4_MOUSE
|
||||||
| NC score | 0.119096 (rank : 36) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9EPK8, Q91XR5, Q9EQZ4, Q9ERZ7, Q9ES76 | Gene names | Trpv4, Trp12, Vrl2, Vroac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (Vanilloid receptor- related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPV2_MOUSE
|
||||||
| NC score | 0.105611 (rank : 37) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WTR1, Q99K71 | Gene names | Trpv2, Grc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Growth factor-regulated calcium channel) (GRC). | |||||
|
NUD9P_HUMAN
|
||||||
| NC score | 0.103483 (rank : 38) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N2Z3 | Gene names | NUDT9P1, C10orf98 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative nucleoside diphosphate-linked moiety X motif 9P1. | |||||
|
PK2L1_HUMAN
|
||||||
| NC score | 0.093806 (rank : 39) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
TRPV2_HUMAN
|
||||||
| NC score | 0.081510 (rank : 40) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5S1, Q9Y670 | Gene names | TRPV2, VRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Vanilloid receptor-like protein 1) (VRL-1). | |||||
|
PK2L2_MOUSE
|
||||||
| NC score | 0.069253 (rank : 41) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L2_HUMAN
|
||||||
| NC score | 0.057296 (rank : 42) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.032488 (rank : 43) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.027366 (rank : 44) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
FANCE_HUMAN
|
||||||
| NC score | 0.025897 (rank : 45) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HB96, Q4ZGH2 | Gene names | FANCE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group E protein (Protein FACE). | |||||
|
CNGA1_HUMAN
|
||||||
| NC score | 0.023862 (rank : 46) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA1_MOUSE
|
||||||
| NC score | 0.020231 (rank : 47) | θ value | 0.0961366 (rank : 31) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CF170_HUMAN
|
||||||
| NC score | 0.017309 (rank : 48) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96NH3, Q6ZMY4, Q6ZUR7, Q8NB47 | Gene names | C6orf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf170. | |||||
|
FRM4B_HUMAN
|
||||||
| NC score | 0.016908 (rank : 49) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2L6, Q8TAI3 | Gene names | FRMD4B, KIAA1013 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1). | |||||
|
UBP37_HUMAN
|
||||||
| NC score | 0.016825 (rank : 50) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
RF1M_HUMAN
|
||||||
| NC score | 0.016038 (rank : 51) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75570, Q5T6Y5, Q8IUQ6 | Gene names | MTRF1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptide chain release factor 1, mitochondrial precursor (MRF-1). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.015445 (rank : 52) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
FRM4B_MOUSE
|
||||||
| NC score | 0.014005 (rank : 53) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q920B0, Q3V1S1, Q9ESP9 | Gene names | Frmd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1) (Golgi- associated band 4.1-like protein) (GOBLIN). | |||||
|
INP4A_MOUSE
|
||||||
| NC score | 0.013718 (rank : 54) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPW0, Q8CBJ8 | Gene names | Inpp4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type I inositol-3,4-bisphosphate 4-phosphatase (EC 3.1.3.66) (Inositol polyphosphate 4-phosphatase type I). | |||||
|
ANLN_HUMAN
|
||||||
| NC score | 0.013480 (rank : 55) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.012908 (rank : 56) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.012876 (rank : 57) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.012409 (rank : 58) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
CCD91_MOUSE
|
||||||
| NC score | 0.012053 (rank : 59) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 699 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D8L5, Q8R3N8, Q9D9E8 | Gene names | Ccdc91, Ggabp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.011976 (rank : 60) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
SLC2B_HUMAN
|
||||||
| NC score | 0.011729 (rank : 61) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NEV8, Q9Y4D6 | Gene names | EXPH5, KIAA0624, SLAC2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Slp homolog lacking C2 domains b (Exophilin-5). | |||||
|
MYPT1_MOUSE
|
||||||
| NC score | 0.011694 (rank : 62) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
DOS_HUMAN
|
||||||
| NC score | 0.011379 (rank : 63) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N350, O43385 | Gene names | DOS, C19orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dos. | |||||
|
MTERF_MOUSE
|
||||||
| NC score | 0.011270 (rank : 64) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CHZ9, Q63ZX5, Q8C7T1 | Gene names | Mterf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor, mitochondrial precursor (mTERF) (Mitochondrial transcription termination factor 1). | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.010958 (rank : 65) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
LRSM1_HUMAN
|
||||||
| NC score | 0.010371 (rank : 66) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.009014 (rank : 67) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
CBPB2_MOUSE
|
||||||
| NC score | 0.009012 (rank : 68) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHH6, Q5EBI3, Q9QZF0 | Gene names | Cpb2, Tafi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase B2 precursor (EC 3.4.17.20) (Carboxypeptidase U) (CPU) (Thrombin-activable fibrinolysis inhibitor) (TAFI) (Carboxypeptidase R) (CPR). | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.008680 (rank : 69) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
NUCL_MOUSE
|
||||||
| NC score | 0.008629 (rank : 70) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
EDN3_MOUSE
|
||||||
| NC score | 0.008548 (rank : 71) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48299, Q543L0 | Gene names | Edn3 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-3 precursor (ET-3) (Preproendothelin-3) (PPET3). | |||||
|
STXB4_HUMAN
|
||||||
| NC score | 0.008342 (rank : 72) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 616 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6ZWJ1, Q8IVZ5 | Gene names | STXBP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
CCD93_MOUSE
|
||||||
| NC score | 0.008195 (rank : 73) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7TQK5, Q3TX53 | Gene names | Ccdc93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 93. | |||||
|
COHA1_HUMAN
|
||||||
| NC score | 0.008120 (rank : 74) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UMD9, Q02802, Q99018, Q9NQK9, Q9UC14 | Gene names | COL17A1, BP180, BPAG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
MYPT1_HUMAN
|
||||||
| NC score | 0.007790 (rank : 75) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.006956 (rank : 76) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
WWTR1_MOUSE
|
||||||
| NC score | 0.006862 (rank : 77) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EPK5, Q99KI4 | Gene names | Wwtr1, Taz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
NCKPL_HUMAN
|
||||||
| NC score | 0.006299 (rank : 78) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55160 | Gene names | NCKAP1L, HEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nck-associated protein 1-like (Membrane-associated protein HEM-1) (Hematopoietic protein 1). | |||||
|
PRGC2_HUMAN
|
||||||
| NC score | 0.005991 (rank : 79) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86YN6, Q86YN3, Q86YN4, Q86YN5, Q8N1N9, Q8TDE4, Q8TDE5 | Gene names | PPARGC1B, PERC, PGC1, PGC1B, PPARGC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta) (PGC-1-related estrogen receptor alpha coactivator) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
BICD1_MOUSE
|
||||||
| NC score | 0.005971 (rank : 80) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
PF21B_HUMAN
|
||||||
| NC score | 0.005435 (rank : 81) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.005371 (rank : 82) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
NMDE1_HUMAN
|
||||||
| NC score | 0.005264 (rank : 83) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12879, O00669 | Gene names | GRIN2A | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A) (hNR2A). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.005242 (rank : 84) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.004656 (rank : 85) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.004044 (rank : 86) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
FA8_MOUSE
|
||||||
| NC score | 0.003533 (rank : 87) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q06194 | Gene names | F8, Cf8, F8c | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | -0.001540 (rank : 88) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ZN142_HUMAN
|
||||||
| NC score | -0.002450 (rank : 89) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||