Please be patient as the page loads
|
ASA3L_HUMAN
|
||||||
| SwissProt Accessions | Q5QJU3, Q569G5, Q5VZR7, Q71RD2 | Gene names | ASAH3L, ACER2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkaline ceramidase 2 (EC 3.5.1.23) (AlkCDase 2) (Acylsphingosine deacylase 3-like) (N-acylsphingosine amidohydrolase 3-like). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ASA3L_HUMAN
|
||||||
| θ value | 2.11095e-166 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5QJU3, Q569G5, Q5VZR7, Q71RD2 | Gene names | ASAH3L, ACER2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkaline ceramidase 2 (EC 3.5.1.23) (AlkCDase 2) (Acylsphingosine deacylase 3-like) (N-acylsphingosine amidohydrolase 3-like). | |||||
|
ASA3L_MOUSE
|
||||||
| θ value | 2.11585e-158 (rank : 2) | NC score | 0.999558 (rank : 2) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VD53, Q6PB92, Q8BUG3 | Gene names | Asah3l, Acer2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkaline ceramidase 2 (EC 3.5.1.23) (AlkCDase 2) (maCER2) (Acylsphingosine deacylase 3-like) (N-acylsphingosine amidohydrolase 3-like) (Cancer related gene-liver 1) (CRG-L1). | |||||
|
ASAH3_HUMAN
|
||||||
| θ value | 2.18062e-54 (rank : 3) | NC score | 0.931863 (rank : 3) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TDN7 | Gene names | ASAH3, ACER1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkaline ceramidase 1 (EC 3.5.1.23) (Alkaline CDase-1) (AlkCDase 1) (Acylsphingosine deacylase 3) (N-acylsphingosine amidohydrolase 3). | |||||
|
ASAH3_MOUSE
|
||||||
| θ value | 7.75577e-52 (rank : 4) | NC score | 0.929263 (rank : 4) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R4X1 | Gene names | Asah3, Acer1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkaline ceramidase 1 (EC 3.5.1.23) (Alkaline CDase-1) (AlkCDase 1) (maCER1) (Acylsphingosine deacylase 3) (N-acylsphingosine amidohydrolase 3). | |||||
|
APHC_HUMAN
|
||||||
| θ value | 6.41864e-14 (rank : 5) | NC score | 0.624842 (rank : 6) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NUN7 | Gene names | PHCA, APHC | |||
|
Domain Architecture |
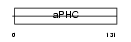
|
|||||
| Description | Alkaline phytoceramidase (EC 3.5.1.-) (aPHC) (Alkaline ceramidase) (Alkaline dihydroceramidase SB89). | |||||
|
APHC_MOUSE
|
||||||
| θ value | 3.18553e-13 (rank : 6) | NC score | 0.627791 (rank : 5) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D099, Q542R2, Q9D0X4, Q9D3J4 | Gene names | Phca, Aphc | |||
|
Domain Architecture |
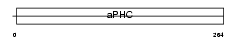
|
|||||
| Description | Alkaline phytoceramidase (EC 3.5.1.-) (aPHC) (Alkaline ceramidase). | |||||
|
TR109_MOUSE
|
||||||
| θ value | 1.06291 (rank : 7) | NC score | 0.017606 (rank : 7) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7M707 | Gene names | Tas2r109, T2r62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 2 member 109 (T2R109) (mT2R62). | |||||
|
NPY5R_HUMAN
|
||||||
| θ value | 6.88961 (rank : 8) | NC score | 0.005544 (rank : 9) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15761, Q92916 | Gene names | NPY5R, NPYR5 | |||
|
Domain Architecture |
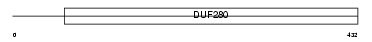
|
|||||
| Description | Neuropeptide Y receptor type 5 (NPY5-R) (NPY-Y5 receptor) (Y5 receptor) (NPYY5). | |||||
|
AT12A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 9) | NC score | 0.008084 (rank : 8) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54707, Q13816, Q13817, Q16734 | Gene names | ATP12A, ATP1AL1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
ASA3L_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.11095e-166 (rank : 1) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5QJU3, Q569G5, Q5VZR7, Q71RD2 | Gene names | ASAH3L, ACER2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkaline ceramidase 2 (EC 3.5.1.23) (AlkCDase 2) (Acylsphingosine deacylase 3-like) (N-acylsphingosine amidohydrolase 3-like). | |||||
|
ASA3L_MOUSE
|
||||||
| NC score | 0.999558 (rank : 2) | θ value | 2.11585e-158 (rank : 2) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VD53, Q6PB92, Q8BUG3 | Gene names | Asah3l, Acer2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkaline ceramidase 2 (EC 3.5.1.23) (AlkCDase 2) (maCER2) (Acylsphingosine deacylase 3-like) (N-acylsphingosine amidohydrolase 3-like) (Cancer related gene-liver 1) (CRG-L1). | |||||
|
ASAH3_HUMAN
|
||||||
| NC score | 0.931863 (rank : 3) | θ value | 2.18062e-54 (rank : 3) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TDN7 | Gene names | ASAH3, ACER1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkaline ceramidase 1 (EC 3.5.1.23) (Alkaline CDase-1) (AlkCDase 1) (Acylsphingosine deacylase 3) (N-acylsphingosine amidohydrolase 3). | |||||
|
ASAH3_MOUSE
|
||||||
| NC score | 0.929263 (rank : 4) | θ value | 7.75577e-52 (rank : 4) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R4X1 | Gene names | Asah3, Acer1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alkaline ceramidase 1 (EC 3.5.1.23) (Alkaline CDase-1) (AlkCDase 1) (maCER1) (Acylsphingosine deacylase 3) (N-acylsphingosine amidohydrolase 3). | |||||
|
APHC_MOUSE
|
||||||
| NC score | 0.627791 (rank : 5) | θ value | 3.18553e-13 (rank : 6) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D099, Q542R2, Q9D0X4, Q9D3J4 | Gene names | Phca, Aphc | |||
|
Domain Architecture |

|
|||||
| Description | Alkaline phytoceramidase (EC 3.5.1.-) (aPHC) (Alkaline ceramidase). | |||||
|
APHC_HUMAN
|
||||||
| NC score | 0.624842 (rank : 6) | θ value | 6.41864e-14 (rank : 5) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NUN7 | Gene names | PHCA, APHC | |||
|
Domain Architecture |

|
|||||
| Description | Alkaline phytoceramidase (EC 3.5.1.-) (aPHC) (Alkaline ceramidase) (Alkaline dihydroceramidase SB89). | |||||
|
TR109_MOUSE
|
||||||
| NC score | 0.017606 (rank : 7) | θ value | 1.06291 (rank : 7) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7M707 | Gene names | Tas2r109, T2r62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 2 member 109 (T2R109) (mT2R62). | |||||
|
AT12A_HUMAN
|
||||||
| NC score | 0.008084 (rank : 8) | θ value | 8.99809 (rank : 9) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54707, Q13816, Q13817, Q16734 | Gene names | ATP12A, ATP1AL1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
NPY5R_HUMAN
|
||||||
| NC score | 0.005544 (rank : 9) | θ value | 6.88961 (rank : 8) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15761, Q92916 | Gene names | NPY5R, NPYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropeptide Y receptor type 5 (NPY5-R) (NPY-Y5 receptor) (Y5 receptor) (NPYY5). | |||||