Please be patient as the page loads
|
PME17_HUMAN
|
||||||
| SwissProt Accessions | P40967, Q12763, Q14448, Q14817, Q16565 | Gene names | SILV, D12S53E, PMEL17 | |||
|
Domain Architecture |

|
|||||
| Description | Melanocyte protein Pmel 17 precursor (Melanocyte lineage-specific antigen GP100) (Melanoma-associated ME20 antigen) (ME20M/ME20S) (ME20- M/ME20-S) (95 kDa melanocyte-specific secreted glycoprotein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PME17_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P40967, Q12763, Q14448, Q14817, Q16565 | Gene names | SILV, D12S53E, PMEL17 | |||
|
Domain Architecture |

|
|||||
| Description | Melanocyte protein Pmel 17 precursor (Melanocyte lineage-specific antigen GP100) (Melanoma-associated ME20 antigen) (ME20M/ME20S) (ME20- M/ME20-S) (95 kDa melanocyte-specific secreted glycoprotein). | |||||
|
PME17_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.952563 (rank : 2) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60696 | Gene names | Silv, D10H12S53E, Pmel17, Si | |||
|
Domain Architecture |

|
|||||
| Description | Melanocyte protein Pmel 17 precursor (Silver locus protein). | |||||
|
GPNMB_HUMAN
|
||||||
| θ value | 4.71619e-41 (rank : 3) | NC score | 0.760006 (rank : 3) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14956, Q6UVX1, Q8N1A1 | Gene names | GPNMB, HGFIN, NMB | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane glycoprotein NMB precursor (Transmembrane glycoprotein HGFIN). | |||||
|
GPNMB_MOUSE
|
||||||
| θ value | 1.98146e-39 (rank : 4) | NC score | 0.753093 (rank : 4) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99P91, Q8BVV9, Q8BXL4, Q9QXA0 | Gene names | Gpnmb, Dchil, Nmb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane glycoprotein NMB precursor (Dendritic cell-associated transmembrane protein) (DC-HIL). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 5) | NC score | 0.170268 (rank : 6) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 6) | NC score | 0.161528 (rank : 8) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
PKD1_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 7) | NC score | 0.098763 (rank : 17) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
MAVS_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 8) | NC score | 0.125955 (rank : 9) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
CK024_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 9) | NC score | 0.169167 (rank : 7) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 10) | NC score | 0.085578 (rank : 25) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 11) | NC score | 0.120404 (rank : 12) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 12) | NC score | 0.067636 (rank : 39) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 13) | NC score | 0.091367 (rank : 20) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 14) | NC score | 0.122873 (rank : 11) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
HCFC1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 15) | NC score | 0.100845 (rank : 16) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
HDAC6_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 16) | NC score | 0.067651 (rank : 38) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UBN7 | Gene names | HDAC6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 17) | NC score | 0.125544 (rank : 10) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
CK024_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 18) | NC score | 0.184949 (rank : 5) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
GP112_HUMAN
|
||||||
| θ value | 0.125558 (rank : 19) | NC score | 0.094454 (rank : 19) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 0.125558 (rank : 20) | NC score | 0.087517 (rank : 23) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
P66A_HUMAN
|
||||||
| θ value | 0.125558 (rank : 21) | NC score | 0.067968 (rank : 35) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86YP4, Q7L3J2, Q96F28, Q9NPU2, Q9NXS1 | Gene names | GATAD2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 alpha (Hp66alpha) (GATA zinc finger domain-containing protein 2A). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 22) | NC score | 0.065624 (rank : 41) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
TIMD1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 23) | NC score | 0.082917 (rank : 27) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96D42, O43656 | Gene names | HAVCR1, TIM1, TIMD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatitis A virus cellular receptor 1 precursor (HAVcr-1) (T cell immunoglobulin and mucin domain-containing protein 1) (TIMD-1) (T cell membrane protein 1) (TIM-1) (TIM). | |||||
|
PRAX_HUMAN
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.061268 (rank : 45) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.105139 (rank : 13) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.063297 (rank : 43) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
ARMX2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.059792 (rank : 52) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
SON_MOUSE
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.102550 (rank : 15) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
ZN628_MOUSE
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.003988 (rank : 113) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 815 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CJ78, Q3U2L5, Q6P5B3 | Gene names | Znf628, Zec, Zfp628 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 628 (Zinc finger protein expressed in embryonal cells and certain adult organs). | |||||
|
ELF2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.025595 (rank : 89) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.080856 (rank : 29) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.103864 (rank : 14) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
YETS2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.075893 (rank : 32) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q3TUF7, Q6PGF8, Q80TI2, Q8CG86 | Gene names | Yeats2, Kiaa1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
K0460_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.064873 (rank : 42) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5VT52, O75048, Q5VT53, Q6MZL4, Q86XD2 | Gene names | KIAA0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
NFASC_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.017485 (rank : 98) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94856, Q5T2F0, Q5T2F1, Q5T2F2, Q5T2F3, Q5T2F4, Q5T2F5, Q5T2F6, Q5T2F7, Q5T2F9, Q5T2G0, Q5W9F8, Q68DH3, Q6ZQV6, Q7Z3K1, Q96HT1, Q96K50 | Gene names | NFASC, KIAA0756 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofascin precursor. | |||||
|
NIPA_MOUSE
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.047392 (rank : 69) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80YV2, Q3TJE6, Q80Z11, Q8BTW5, Q8CI56, Q8R3U7 | Gene names | Zc3hc1, Nipa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear-interacting partner of ALK (Nuclear-interacting partner of anaplastic lymphoma kinase) (mNIPA) (Zinc finger C3HC-type protein 1). | |||||
|
PK1L1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.086134 (rank : 24) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
USH2A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.022754 (rank : 93) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75445, Q5VVM9, Q6S362, Q9NS27 | Gene names | USH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein) (Usher syndrome type IIa protein). | |||||
|
HCFC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.097559 (rank : 18) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
PCDH9_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.006691 (rank : 108) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HC56 | Gene names | PCDH9 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-9 precursor. | |||||
|
SON_HUMAN
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.090820 (rank : 21) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
RBL2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.025233 (rank : 90) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64700 | Gene names | Rbl2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-like protein 2 (130 kDa retinoblastoma-associated protein) (PRB2) (P130) (RBR-2). | |||||
|
TACC2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.028635 (rank : 86) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
ASM3A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.024132 (rank : 91) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70158 | Gene names | Smpdl3a, Asml3a | |||
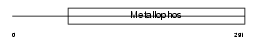
|
Domain Architecture |
|
|||||
| Description | Acid sphingomyelinase-like phosphodiesterase 3a precursor (EC 3.1.4.-) (ASM-like phosphodiesterase 3a). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.055162 (rank : 58) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.019749 (rank : 95) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
IF4G3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.031533 (rank : 82) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.030593 (rank : 83) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
MYBPH_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.017673 (rank : 97) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70402 | Gene names | Mybph | |||
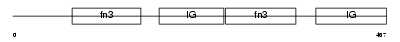
|
Domain Architecture |
|
|||||
| Description | Myosin-binding protein H (MyBP-H) (H-protein). | |||||
|
SPR2I_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.059922 (rank : 51) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70560 | Gene names | Sprr2i | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2I. | |||||
|
TRAK1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.019141 (rank : 96) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
CD45_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.028812 (rank : 85) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P08575, Q16614, Q9H0Y6 | Gene names | PTPRC, CD45 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (CD45 antigen) (T200). | |||||
|
HPT_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.007748 (rank : 106) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61646 | Gene names | Hp | |||
|
Domain Architecture |

|
|||||
| Description | Haptoglobin precursor [Contains: Haptoglobin alpha chain; Haptoglobin beta chain]. | |||||
|
INP5E_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.026389 (rank : 88) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JII1, Q3TCC9 | Gene names | Inpp5e | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
KLF7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.004582 (rank : 112) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 686 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75840 | Gene names | KLF7, UKLF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 7 (Ubiquitous krueppel-like factor). | |||||
|
TCF8_MOUSE
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.005969 (rank : 109) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
CSF1R_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.002280 (rank : 115) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 943 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P07333, Q6LDW5, Q6LDY4 | Gene names | CSF1R, FMS | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 receptor precursor (EC 2.7.10.1) (CSF-1-R) (Fms proto-oncogene) (c-fms) (CD115 antigen). | |||||
|
GMEB2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.023175 (rank : 92) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P58929, Q6PCY0 | Gene names | Gmeb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucocorticoid modulatory element-binding protein 2 (GMEB-2). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.046741 (rank : 72) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.078841 (rank : 30) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
NELFA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.047004 (rank : 71) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.047004 (rank : 70) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PTPRN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.012537 (rank : 104) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
SPR2E_MOUSE
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.049879 (rank : 68) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70556 | Gene names | Sprr2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E. | |||||
|
SPR2F_MOUSE
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.050144 (rank : 67) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70557 | Gene names | Sprr2f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F. | |||||
|
TIEG3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.007596 (rank : 107) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K1S5, Q8BI37 | Gene names | Tieg3, Tieg2b | |||
|
Domain Architecture |

|
|||||
| Description | Transforming growth factor-beta-inducible early growth response protein 3 (TGFB-inducible early growth response protein 3) (TIEG-3) (TGFB-inducible early growth response protein 2b). | |||||
|
K0460_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.068354 (rank : 34) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
MEOX1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.004708 (rank : 111) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P32442 | Gene names | Meox1, Mox-1, Mox1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein MOX-1 (Mesenchyme homeobox 1). | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.041917 (rank : 77) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
TESK1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.002907 (rank : 114) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15569 | Gene names | TESK1 | |||
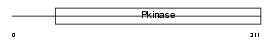
|
Domain Architecture |
|
|||||
| Description | Dual specificity testis-specific protein kinase 1 (EC 2.7.12.1) (Testicular protein kinase 1). | |||||
|
DAF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.021604 (rank : 94) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61475, P97732, Q61397 | Gene names | Cd55a, Daf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor, GPI-anchored precursor (DAF-GPI) (CD55 antigen). | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.013262 (rank : 102) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
NUP62_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.059242 (rank : 54) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q63850, Q99JN7 | Gene names | Nup62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
TMIGD_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.013539 (rank : 101) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6UXZ0, Q6ZMC6 | Gene names | TMIGD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and immunoglobulin domain-containing protein precursor. | |||||
|
VP13B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.026952 (rank : 87) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z7G8, Q709C6, Q709C7, Q7Z7G4, Q7Z7G5, Q7Z7G6, Q7Z7G7, Q8NB77, Q9NWV1, Q9Y4E7 | Gene names | VPS13B, CHS1, COH1, KIAA0532 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 13B (Cohen syndrome protein 1). | |||||
|
YETS2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.065848 (rank : 40) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.042879 (rank : 76) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.038907 (rank : 78) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
DYNA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.013093 (rank : 103) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.072636 (rank : 33) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.083602 (rank : 26) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
P66A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.045958 (rank : 73) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHY6, Q8BTQ2, Q8VEC9 | Gene names | Gatad2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 alpha (GATA zinc finger domain- containing protein 2A). | |||||
|
SDC3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.067798 (rank : 37) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q64519, Q3UDD9, Q6ZQA4, Q7TQD4 | Gene names | Sdc3, Kiaa0468 | |||
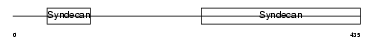
|
Domain Architecture |
|
|||||
| Description | Syndecan-3 precursor (SYND3). | |||||
|
SF3B1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.044201 (rank : 74) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.044173 (rank : 75) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
TACC3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.017292 (rank : 99) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JJ11, Q9WVK9 | Gene names | Tacc3, Aint | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ARNT-interacting protein). | |||||
|
ZN236_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.001981 (rank : 116) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UL36, Q9UL37 | Gene names | ZNF236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 236. | |||||
|
ALG12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.013599 (rank : 100) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BV10, Q8NG10, Q96AA4 | Gene names | ALG12 | |||
|
Domain Architecture |

|
|||||
| Description | Dolichyl-P-Man:Man(7)GlcNAc(2)-PP-dolichyl-alpha-1,6- mannosyltransferase (EC 2.4.1.-) (Mannosyltransferase ALG12 homolog) (hALG12) (Membrane protein SB87). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.035700 (rank : 79) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.035683 (rank : 80) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
FOXO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.005833 (rank : 110) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12778, O43523, Q5VYC7, Q6NSK6 | Gene names | FOXO1A, FKHR | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein O1A (Forkhead in rhabdomyosarcoma). | |||||
|
RD23B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.029116 (rank : 84) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P54727, Q8WUB0 | Gene names | RAD23B | |||
|
Domain Architecture |
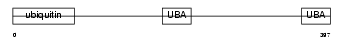
|
|||||
| Description | UV excision repair protein RAD23 homolog B (hHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
SOCS3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.010440 (rank : 105) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35718, P97803, Q3U7X5 | Gene names | Socs3, Cis3, Cish3 | |||
|
Domain Architecture |
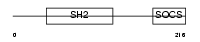
|
|||||
| Description | Suppressor of cytokine signaling 3 (SOCS-3) (Cytokine-inducible SH2 protein 3) (CIS-3) (Protein EF-10). | |||||
|
SPR2D_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.031578 (rank : 81) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70555 | Gene names | Sprr2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D. | |||||
|
TESK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.001716 (rank : 117) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70146, O70147 | Gene names | Tesk1 | |||
|
Domain Architecture |
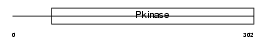
|
|||||
| Description | Dual specificity testis-specific protein kinase 1 (EC 2.7.12.1) (Testicular protein kinase 1). | |||||
|
CO002_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.051295 (rank : 65) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
K1683_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.053490 (rank : 60) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
LEUK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.090137 (rank : 22) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P16150 | Gene names | SPN, CD43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Galactoglycoprotein) (GALGP) (CD43 antigen). | |||||
|
LEUK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.060354 (rank : 47) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15702 | Gene names | Spn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Ly- 48) (B cell differentiation antigen LP-3) (CD43 antigen). | |||||
|
LR37A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.052843 (rank : 61) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
MAP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.058044 (rank : 56) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MAVS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.060058 (rank : 49) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VCF0 | Gene names | Mavs, Ips1, Visa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif). | |||||
|
MUCEN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.052511 (rank : 63) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R0H2, Q78KL2, Q9DCN9, Q9ULC1, Q9Z2I1 | Gene names | Emcn, Muc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endomucin precursor (Endomucin-1/2). | |||||
|
NFRKB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.056905 (rank : 57) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
NU214_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.062933 (rank : 44) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
PARM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.060032 (rank : 50) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
PODXL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.077351 (rank : 31) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00592 | Gene names | PODXL, PCLP, PCLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.058672 (rank : 55) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SDC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.052733 (rank : 62) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75056, Q5T1Z6, Q5T1Z7, Q96CT3, Q96PR8 | Gene names | SDC3, KIAA0468 | |||
|
Domain Architecture |
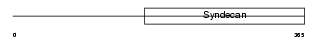
|
|||||
| Description | Syndecan-3 (SYND3). | |||||
|
SELPL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.059661 (rank : 53) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14242, Q12775 | Gene names | SELPLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand) (CD162 antigen). | |||||
|
SPRR3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.052086 (rank : 64) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O09116 | Gene names | Sprr3 | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.067964 (rank : 36) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
TECTA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.050351 (rank : 66) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75443 | Gene names | TECTA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
TM108_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.053493 (rank : 59) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
VWF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.060214 (rank : 48) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
VWF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.061001 (rank : 46) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
ZAN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.082803 (rank : 28) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
PME17_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P40967, Q12763, Q14448, Q14817, Q16565 | Gene names | SILV, D12S53E, PMEL17 | |||
|
Domain Architecture |

|
|||||
| Description | Melanocyte protein Pmel 17 precursor (Melanocyte lineage-specific antigen GP100) (Melanoma-associated ME20 antigen) (ME20M/ME20S) (ME20- M/ME20-S) (95 kDa melanocyte-specific secreted glycoprotein). | |||||
|
PME17_MOUSE
|
||||||
| NC score | 0.952563 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60696 | Gene names | Silv, D10H12S53E, Pmel17, Si | |||
|
Domain Architecture |

|
|||||
| Description | Melanocyte protein Pmel 17 precursor (Silver locus protein). | |||||
|
GPNMB_HUMAN
|
||||||
| NC score | 0.760006 (rank : 3) | θ value | 4.71619e-41 (rank : 3) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14956, Q6UVX1, Q8N1A1 | Gene names | GPNMB, HGFIN, NMB | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane glycoprotein NMB precursor (Transmembrane glycoprotein HGFIN). | |||||
|
GPNMB_MOUSE
|
||||||
| NC score | 0.753093 (rank : 4) | θ value | 1.98146e-39 (rank : 4) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99P91, Q8BVV9, Q8BXL4, Q9QXA0 | Gene names | Gpnmb, Dchil, Nmb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane glycoprotein NMB precursor (Dendritic cell-associated transmembrane protein) (DC-HIL). | |||||
|
CK024_MOUSE
|
||||||
| NC score | 0.184949 (rank : 5) | θ value | 0.0961366 (rank : 18) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.170268 (rank : 6) | θ value | 2.36244e-08 (rank : 5) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.169167 (rank : 7) | θ value | 0.00869519 (rank : 9) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.161528 (rank : 8) | θ value | 4.1701e-05 (rank : 6) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
MAVS_HUMAN
|
||||||
| NC score | 0.125955 (rank : 9) | θ value | 0.00665767 (rank : 8) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.125544 (rank : 10) | θ value | 0.0736092 (rank : 17) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.122873 (rank : 11) | θ value | 0.0330416 (rank : 14) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.120404 (rank : 12) | θ value | 0.0193708 (rank : 11) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.105139 (rank : 13) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.103864 (rank : 14) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.102550 (rank : 15) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
HCFC1_MOUSE
|
||||||
| NC score | 0.100845 (rank : 16) | θ value | 0.0736092 (rank : 15) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
PKD1_HUMAN
|
||||||
| NC score | 0.098763 (rank : 17) | θ value | 0.00509761 (rank : 7) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
HCFC1_HUMAN
|
||||||
| NC score | 0.097559 (rank : 18) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.094454 (rank : 19) | θ value | 0.125558 (rank : 19) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.091367 (rank : 20) | θ value | 0.0330416 (rank : 13) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.090820 (rank : 21) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
LEUK_HUMAN
|
||||||
| NC score | 0.090137 (rank : 22) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P16150 | Gene names | SPN, CD43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Galactoglycoprotein) (GALGP) (CD43 antigen). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.087517 (rank : 23) | θ value | 0.125558 (rank : 20) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
PK1L1_HUMAN
|
||||||
| NC score | 0.086134 (rank : 24) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.085578 (rank : 25) | θ value | 0.0113563 (rank : 10) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.083602 (rank : 26) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
TIMD1_HUMAN
|
||||||
| NC score | 0.082917 (rank : 27) | θ value | 0.125558 (rank : 23) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96D42, O43656 | Gene names | HAVCR1, TIM1, TIMD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatitis A virus cellular receptor 1 precursor (HAVcr-1) (T cell immunoglobulin and mucin domain-containing protein 1) (TIMD-1) (T cell membrane protein 1) (TIM-1) (TIM). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.082803 (rank : 28) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.080856 (rank : 29) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.078841 (rank : 30) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
PODXL_HUMAN
|
||||||
| NC score | 0.077351 (rank : 31) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00592 | Gene names | PODXL, PCLP, PCLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
YETS2_MOUSE
|
||||||
| NC score | 0.075893 (rank : 32) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q3TUF7, Q6PGF8, Q80TI2, Q8CG86 | Gene names | Yeats2, Kiaa1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.072636 (rank : 33) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
K0460_MOUSE
|
||||||
| NC score | 0.068354 (rank : 34) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
P66A_HUMAN
|
||||||
| NC score | 0.067968 (rank : 35) | θ value | 0.125558 (rank : 21) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86YP4, Q7L3J2, Q96F28, Q9NPU2, Q9NXS1 | Gene names | GATAD2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 alpha (Hp66alpha) (GATA zinc finger domain-containing protein 2A). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.067964 (rank : 36) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
SDC3_MOUSE
|
||||||
| NC score | 0.067798 (rank : 37) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q64519, Q3UDD9, Q6ZQA4, Q7TQD4 | Gene names | Sdc3, Kiaa0468 | |||
|
Domain Architecture |

|
|||||
| Description | Syndecan-3 precursor (SYND3). | |||||
|
HDAC6_HUMAN
|
||||||
| NC score | 0.067651 (rank : 38) | θ value | 0.0736092 (rank : 16) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UBN7 | Gene names | HDAC6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6). | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.067636 (rank : 39) | θ value | 0.0252991 (rank : 12) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
YETS2_HUMAN
|
||||||
| NC score | 0.065848 (rank : 40) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.065624 (rank : 41) | θ value | 0.125558 (rank : 22) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
K0460_HUMAN
|
||||||
| NC score | 0.064873 (rank : 42) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5VT52, O75048, Q5VT53, Q6MZL4, Q86XD2 | Gene names | KIAA0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.063297 (rank : 43) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.062933 (rank : 44) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
PRAX_HUMAN
|
||||||
| NC score | 0.061268 (rank : 45) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
VWF_MOUSE
|
||||||
| NC score | 0.061001 (rank : 46) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
LEUK_MOUSE
|
||||||
| NC score | 0.060354 (rank : 47) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15702 | Gene names | Spn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Ly- 48) (B cell differentiation antigen LP-3) (CD43 antigen). | |||||
|
VWF_HUMAN
|
||||||
| NC score | 0.060214 (rank : 48) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
MAVS_MOUSE
|
||||||
| NC score | 0.060058 (rank : 49) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VCF0 | Gene names | Mavs, Ips1, Visa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif). | |||||
|
PARM1_MOUSE
|
||||||
| NC score | 0.060032 (rank : 50) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
SPR2I_MOUSE
|
||||||
| NC score | 0.059922 (rank : 51) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70560 | Gene names | Sprr2i | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2I. | |||||
|
ARMX2_MOUSE
|
||||||
| NC score | 0.059792 (rank : 52) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
SELPL_HUMAN
|
||||||
| NC score | 0.059661 (rank : 53) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14242, Q12775 | Gene names | SELPLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand) (CD162 antigen). | |||||
|
NUP62_MOUSE
|
||||||
| NC score | 0.059242 (rank : 54) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q63850, Q99JN7 | Gene names | Nup62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.058672 (rank : 55) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.058044 (rank : 56) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
NFRKB_MOUSE
|
||||||
| NC score | 0.056905 (rank : 57) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.055162 (rank : 58) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
TM108_MOUSE
|
||||||
| NC score | 0.053493 (rank : 59) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
K1683_HUMAN
|
||||||
| NC score | 0.053490 (rank : 60) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.052843 (rank : 61) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
SDC3_HUMAN
|
||||||
| NC score | 0.052733 (rank : 62) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75056, Q5T1Z6, Q5T1Z7, Q96CT3, Q96PR8 | Gene names | SDC3, KIAA0468 | |||
|
Domain Architecture |

|
|||||
| Description | Syndecan-3 (SYND3). | |||||
|
MUCEN_MOUSE
|
||||||
| NC score | 0.052511 (rank : 63) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R0H2, Q78KL2, Q9DCN9, Q9ULC1, Q9Z2I1 | Gene names | Emcn, Muc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endomucin precursor (Endomucin-1/2). | |||||
|
SPRR3_MOUSE
|
||||||
| NC score | 0.052086 (rank : 64) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O09116 | Gene names | Sprr3 | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta). | |||||
|
CO002_HUMAN
|
||||||
| NC score | 0.051295 (rank : 65) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
TECTA_HUMAN
|
||||||
| NC score | 0.050351 (rank : 66) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75443 | Gene names | TECTA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
SPR2F_MOUSE
|
||||||
| NC score | 0.050144 (rank : 67) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70557 | Gene names | Sprr2f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F. | |||||
|
SPR2E_MOUSE
|
||||||
| NC score | 0.049879 (rank : 68) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70556 | Gene names | Sprr2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E. | |||||
|
NIPA_MOUSE
|
||||||
| NC score | 0.047392 (rank : 69) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80YV2, Q3TJE6, Q80Z11, Q8BTW5, Q8CI56, Q8R3U7 | Gene names | Zc3hc1, Nipa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear-interacting partner of ALK (Nuclear-interacting partner of anaplastic lymphoma kinase) (mNIPA) (Zinc finger C3HC-type protein 1). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.047004 (rank : 70) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
NELFA_MOUSE
|
||||||
| NC score | 0.047004 (rank : 71) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.046741 (rank : 72) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
P66A_MOUSE
|
||||||
| NC score | 0.045958 (rank : 73) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHY6, Q8BTQ2, Q8VEC9 | Gene names | Gatad2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 alpha (GATA zinc finger domain- containing protein 2A). | |||||
|
SF3B1_HUMAN
|
||||||
| NC score | 0.044201 (rank : 74) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| NC score | 0.044173 (rank : 75) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.042879 (rank : 76) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.041917 (rank : 77) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.038907 (rank : 78) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.035700 (rank : 79) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.035683 (rank : 80) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
SPR2D_MOUSE
|
||||||
| NC score | 0.031578 (rank : 81) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70555 | Gene names | Sprr2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D. | |||||
|
IF4G3_HUMAN
|
||||||
| NC score | 0.031533 (rank : 82) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.030593 (rank : 83) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
RD23B_HUMAN
|
||||||
| NC score | 0.029116 (rank : 84) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P54727, Q8WUB0 | Gene names | RAD23B | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (hHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
CD45_HUMAN
|
||||||
| NC score | 0.028812 (rank : 85) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P08575, Q16614, Q9H0Y6 | Gene names | PTPRC, CD45 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (CD45 antigen) (T200). | |||||
|
TACC2_HUMAN
|
||||||
| NC score | 0.028635 (rank : 86) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
VP13B_HUMAN
|
||||||
| NC score | 0.026952 (rank : 87) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z7G8, Q709C6, Q709C7, Q7Z7G4, Q7Z7G5, Q7Z7G6, Q7Z7G7, Q8NB77, Q9NWV1, Q9Y4E7 | Gene names | VPS13B, CHS1, COH1, KIAA0532 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 13B (Cohen syndrome protein 1). | |||||
|
INP5E_MOUSE
|
||||||
| NC score | 0.026389 (rank : 88) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JII1, Q3TCC9 | Gene names | Inpp5e | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
ELF2_HUMAN
|
||||||
| NC score | 0.025595 (rank : 89) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
RBL2_MOUSE
|
||||||
| NC score | 0.025233 (rank : 90) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64700 | Gene names | Rbl2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-like protein 2 (130 kDa retinoblastoma-associated protein) (PRB2) (P130) (RBR-2). | |||||
|
ASM3A_MOUSE
|
||||||
| NC score | 0.024132 (rank : 91) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70158 | Gene names | Smpdl3a, Asml3a | |||
|
Domain Architecture |

|
|||||
| Description | Acid sphingomyelinase-like phosphodiesterase 3a precursor (EC 3.1.4.-) (ASM-like phosphodiesterase 3a). | |||||
|
GMEB2_MOUSE
|
||||||
| NC score | 0.023175 (rank : 92) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P58929, Q6PCY0 | Gene names | Gmeb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucocorticoid modulatory element-binding protein 2 (GMEB-2). | |||||
|
USH2A_HUMAN
|
||||||
| NC score | 0.022754 (rank : 93) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75445, Q5VVM9, Q6S362, Q9NS27 | Gene names | USH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein) (Usher syndrome type IIa protein). | |||||
|
DAF1_MOUSE
|
||||||
| NC score | 0.021604 (rank : 94) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61475, P97732, Q61397 | Gene names | Cd55a, Daf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor, GPI-anchored precursor (DAF-GPI) (CD55 antigen). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.019749 (rank : 95) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
TRAK1_HUMAN
|
||||||
| NC score | 0.019141 (rank : 96) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
MYBPH_MOUSE
|
||||||
| NC score | 0.017673 (rank : 97) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70402 | Gene names | Mybph | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-binding protein H (MyBP-H) (H-protein). | |||||
|
NFASC_HUMAN
|
||||||
| NC score | 0.017485 (rank : 98) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94856, Q5T2F0, Q5T2F1, Q5T2F2, Q5T2F3, Q5T2F4, Q5T2F5, Q5T2F6, Q5T2F7, Q5T2F9, Q5T2G0, Q5W9F8, Q68DH3, Q6ZQV6, Q7Z3K1, Q96HT1, Q96K50 | Gene names | NFASC, KIAA0756 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofascin precursor. | |||||
|
TACC3_MOUSE
|
||||||
| NC score | 0.017292 (rank : 99) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JJ11, Q9WVK9 | Gene names | Tacc3, Aint | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ARNT-interacting protein). | |||||
|
ALG12_HUMAN
|
||||||
| NC score | 0.013599 (rank : 100) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BV10, Q8NG10, Q96AA4 | Gene names | ALG12 | |||
|
Domain Architecture |

|
|||||
| Description | Dolichyl-P-Man:Man(7)GlcNAc(2)-PP-dolichyl-alpha-1,6- mannosyltransferase (EC 2.4.1.-) (Mannosyltransferase ALG12 homolog) (hALG12) (Membrane protein SB87). | |||||
|
TMIGD_HUMAN
|
||||||
| NC score | 0.013539 (rank : 101) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6UXZ0, Q6ZMC6 | Gene names | TMIGD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and immunoglobulin domain-containing protein precursor. | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.013262 (rank : 102) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
DYNA_MOUSE
|
||||||
| NC score | 0.013093 (rank : 103) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
PTPRN_MOUSE
|
||||||
| NC score | 0.012537 (rank : 104) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
SOCS3_MOUSE
|
||||||
| NC score | 0.010440 (rank : 105) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35718, P97803, Q3U7X5 | Gene names | Socs3, Cis3, Cish3 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 3 (SOCS-3) (Cytokine-inducible SH2 protein 3) (CIS-3) (Protein EF-10). | |||||
|
HPT_MOUSE
|
||||||
| NC score | 0.007748 (rank : 106) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61646 | Gene names | Hp | |||
|
Domain Architecture |

|
|||||
| Description | Haptoglobin precursor [Contains: Haptoglobin alpha chain; Haptoglobin beta chain]. | |||||
|
TIEG3_MOUSE
|
||||||
| NC score | 0.007596 (rank : 107) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K1S5, Q8BI37 | Gene names | Tieg3, Tieg2b | |||
|
Domain Architecture |

|
|||||
| Description | Transforming growth factor-beta-inducible early growth response protein 3 (TGFB-inducible early growth response protein 3) (TIEG-3) (TGFB-inducible early growth response protein 2b). | |||||
|
PCDH9_HUMAN
|
||||||
| NC score | 0.006691 (rank : 108) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HC56 | Gene names | PCDH9 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-9 precursor. | |||||
|
TCF8_MOUSE
|
||||||
| NC score | 0.005969 (rank : 109) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
FOXO1_HUMAN
|
||||||
| NC score | 0.005833 (rank : 110) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12778, O43523, Q5VYC7, Q6NSK6 | Gene names | FOXO1A, FKHR | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein O1A (Forkhead in rhabdomyosarcoma). | |||||
|
MEOX1_MOUSE
|
||||||
| NC score | 0.004708 (rank : 111) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P32442 | Gene names | Meox1, Mox-1, Mox1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein MOX-1 (Mesenchyme homeobox 1). | |||||
|
KLF7_HUMAN
|
||||||
| NC score | 0.004582 (rank : 112) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 686 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75840 | Gene names | KLF7, UKLF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 7 (Ubiquitous krueppel-like factor). | |||||
|
ZN628_MOUSE
|
||||||
| NC score | 0.003988 (rank : 113) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 815 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CJ78, Q3U2L5, Q6P5B3 | Gene names | Znf628, Zec, Zfp628 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 628 (Zinc finger protein expressed in embryonal cells and certain adult organs). | |||||
|
TESK1_HUMAN
|
||||||
| NC score | 0.002907 (rank : 114) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15569 | Gene names | TESK1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity testis-specific protein kinase 1 (EC 2.7.12.1) (Testicular protein kinase 1). | |||||
|
CSF1R_HUMAN
|
||||||
| NC score | 0.002280 (rank : 115) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 943 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P07333, Q6LDW5, Q6LDY4 | Gene names | CSF1R, FMS | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 receptor precursor (EC 2.7.10.1) (CSF-1-R) (Fms proto-oncogene) (c-fms) (CD115 antigen). | |||||
|
ZN236_HUMAN
|
||||||
| NC score | 0.001981 (rank : 116) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UL36, Q9UL37 | Gene names | ZNF236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 236. | |||||
|
TESK1_MOUSE
|
||||||
| NC score | 0.001716 (rank : 117) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70146, O70147 | Gene names | Tesk1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity testis-specific protein kinase 1 (EC 2.7.12.1) (Testicular protein kinase 1). | |||||