Please be patient as the page loads
|
FRS3_HUMAN
|
||||||
| SwissProt Accessions | O43559, Q5T3D5 | Gene names | FRS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (Suc1-associated neurotrophic factor target 2) (SNT-2) (FGFR signaling adaptor SNT2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FRS3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O43559, Q5T3D5 | Gene names | FRS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (Suc1-associated neurotrophic factor target 2) (SNT-2) (FGFR signaling adaptor SNT2). | |||||
|
FRS3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.949636 (rank : 4) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91WJ0 | Gene names | Frs3, Frs2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (FRS2-beta). | |||||
|
FRS2_HUMAN
|
||||||
| θ value | 4.92331e-123 (rank : 3) | NC score | 0.952806 (rank : 2) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WU20, O43558, Q7LDQ6 | Gene names | FRS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 2 (FGFR substrate 2) (Suc1-associated neurotrophic factor target 1) (SNT-1) (FGFR signaling adaptor SNT). | |||||
|
FRS2_MOUSE
|
||||||
| θ value | 8.39787e-123 (rank : 4) | NC score | 0.951216 (rank : 3) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C180 | Gene names | Frs2, Frs2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 2 (FGFR substrate 2) (Suc1-associated neurotrophic factor target 1) (SNT-1) (FGFR signaling adaptor SNT) (FRS2 alpha). | |||||
|
DOK1_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 5) | NC score | 0.519437 (rank : 6) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97465, Q9R213 | Gene names | Dok1, Dok | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 1 (Downstream of tyrosine kinase 1) (p62(dok)). | |||||
|
DOK1_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 6) | NC score | 0.519535 (rank : 5) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99704, O43204, Q9UHG6 | Gene names | DOK1 | |||
|
Domain Architecture |
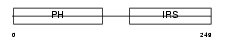
|
|||||
| Description | Docking protein 1 (Downstream of tyrosine kinase 1) (p62(dok)) (pp62). | |||||
|
DOK6_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 7) | NC score | 0.479325 (rank : 11) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PKX4, Q4V9S3, Q8WUZ8, Q96HI2, Q96LU2 | Gene names | DOK6, DOK5L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 6 (Downstream of tyrosine kinase 6). | |||||
|
DOK2_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 8) | NC score | 0.480451 (rank : 10) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O70469, O70272, Q99KL1 | Gene names | Dok2, Frip | |||
|
Domain Architecture |
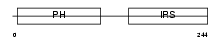
|
|||||
| Description | Docking protein 2 (Downstream of tyrosine kinase 2) (p56(dok-2)) (Dok- related protein) (Dok-R) (IL-four receptor-interacting protein) (FRIP). | |||||
|
DOK3_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 9) | NC score | 0.516986 (rank : 7) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QZK7 | Gene names | Dok3, Dokl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 3 (Downstream of tyrosine kinase 3) (P62(dok)-like protein) (DOK-L). | |||||
|
DOK3_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 10) | NC score | 0.500677 (rank : 9) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7L591, Q8N864, Q9BQB3, Q9H666 | Gene names | DOK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 3 (Downstream of tyrosine kinase 3). | |||||
|
DOK2_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 11) | NC score | 0.508692 (rank : 8) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60496 | Gene names | DOK2 | |||
|
Domain Architecture |
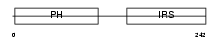
|
|||||
| Description | Docking protein 2 (Downstream of tyrosine kinase 2) (p56(dok-2)). | |||||
|
DOK4_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 12) | NC score | 0.452519 (rank : 14) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TEW6, O75209, Q9BTP2, Q9NVV3 | Gene names | DOK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 4 (Downstream of tyrosine kinase 4). | |||||
|
DOK4_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 13) | NC score | 0.450422 (rank : 15) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99KE3, Q3U0X3 | Gene names | Dok4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 4 (Downstream of tyrosine kinase 4). | |||||
|
DOK5_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 14) | NC score | 0.454897 (rank : 12) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9P104, Q5T7Y0, Q5TE53, Q8TEW7, Q96H13, Q9BZ24, Q9NQF4, Q9Y411 | Gene names | DOK5, C20orf180 | |||
|
Domain Architecture |
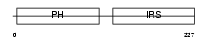
|
|||||
| Description | Docking protein 5 (Downstream of tyrosine kinase 5) (IRS6) (Protein dok-5). | |||||
|
DOK5_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 15) | NC score | 0.453841 (rank : 13) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91ZM9, Q8BRI3, Q9CSM6 | Gene names | Dok5 | |||
|
Domain Architecture |
|
|||||
| Description | Docking protein 5 (Downstream of tyrosine kinase 5) (Protein dok-5). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 16) | NC score | 0.045414 (rank : 29) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 17) | NC score | 0.068763 (rank : 20) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 18) | NC score | 0.080262 (rank : 19) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
DOK7_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 19) | NC score | 0.236817 (rank : 16) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q18PE1, Q6P6A6, Q86XG5, Q8N2J3, Q8NBC1 | Gene names | DOK7, C4orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 20) | NC score | 0.058889 (rank : 23) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
DOK7_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 21) | NC score | 0.227030 (rank : 17) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 22) | NC score | 0.050395 (rank : 26) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 23) | NC score | 0.059965 (rank : 22) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
PRP1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.033727 (rank : 34) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
CLCN1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.023608 (rank : 46) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64347 | Gene names | Clcn1, Clc1 | |||
|
Domain Architecture |
|
|||||
| Description | Chloride channel protein, skeletal muscle (Chloride channel protein 1) (ClC-1). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.036444 (rank : 31) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
CBL_MOUSE
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.029906 (rank : 38) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P22682, Q8CEA1 | Gene names | Cbl | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.051411 (rank : 25) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
IP3KA_MOUSE
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.032266 (rank : 36) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R071 | Gene names | Itpka | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol-trisphosphate 3-kinase A (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase A) (IP3K A) (IP3 3-kinase A). | |||||
|
SAPS1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.029946 (rank : 37) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.034386 (rank : 33) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
ADA19_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.014096 (rank : 59) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H013, Q9BZL5, Q9UHP2 | Gene names | ADAM19, MLTNB | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 19) (Meltrin beta) (Metalloprotease and disintegrin dentritic antigen marker) (MADDAM). | |||||
|
CLMN_MOUSE
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.014171 (rank : 58) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C5W0, Q91V71, Q91XT7, Q91XT8, Q91XU9 | Gene names | Clmn | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
HNRPG_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.022518 (rank : 49) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P38159, Q5JQ67, Q969R3 | Gene names | RBMX, HNRPG, RBMXP1 | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein G (hnRNP G) (RNA-binding motif protein, X chromosome) (Glycoprotein p43). | |||||
|
KCNH2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.022513 (rank : 50) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
NED4L_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.023337 (rank : 47) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NED4L_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.023744 (rank : 45) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
PER1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.021478 (rank : 51) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.029684 (rank : 40) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.029594 (rank : 41) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TAOK2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.001406 (rank : 77) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1373 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZQ29, Q7TSS8 | Gene names | Taok2, Kiaa0881 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2). | |||||
|
PRPE_HUMAN
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.089188 (rank : 18) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P02811 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide P-E (IB-9). | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.014267 (rank : 57) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.032539 (rank : 35) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
WASP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.047062 (rank : 28) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
MAP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.029887 (rank : 39) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.022822 (rank : 48) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CSKI2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.015839 (rank : 55) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
NEB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.011640 (rank : 64) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
PRP5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.040064 (rank : 30) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
ARP5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.011687 (rank : 63) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H9F9, Q86WF7, Q8IUY5, Q8N724, Q9BRN0, Q9BVB7 | Gene names | ACTR5, ARP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-related protein 5 (hARP5) (Sarcoma antigen NY-SAR-16). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.007945 (rank : 69) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.025837 (rank : 42) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
NEDD4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.016990 (rank : 54) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.007476 (rank : 70) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.068745 (rank : 21) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
COIA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.007370 (rank : 71) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
CSKI2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.020958 (rank : 52) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
HNRPG_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.024167 (rank : 44) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35479 | Gene names | Rbmx, Hnrnpg, Hnrpg, Rbmxp1, Rbmxrt | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein G (hnRNP G) (RNA-binding motif protein, X chromosome). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.012985 (rank : 60) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
PSD4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.012400 (rank : 62) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NDX1, O95621, Q4ZG34, Q6GPH8, Q8IYP4 | Gene names | PSD4, EFA6B, TIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6B) (Telomeric of interleukin-1 cluster protein). | |||||
|
SIRT6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.012829 (rank : 61) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N6T7, O75291, Q6IAF5, Q6PK99, Q8NCD2, Q9BSI5, Q9BWP3, Q9NRC7, Q9UQD1 | Gene names | SIRT6, SIR2L6 | |||
|
Domain Architecture |
|
|||||
| Description | Mono-ADP-ribosyltransferase sirtuin-6 (EC 2.4.2.31) (SIR2-like protein 6). | |||||
|
ACK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.002660 (rank : 75) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1140 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q07912, Q8N6U7, Q96H59 | Gene names | TNK2, ACK1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Tyrosine kinase non- receptor protein 2). | |||||
|
B3A2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.006703 (rank : 72) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04920, Q45EY5, Q969L3 | Gene names | SLC4A2, AE2, EPB3L1, HKB3, MPB3L | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 2 (Non-erythroid band 3-like protein) (AE2 anion exchanger) (Solute carrier family 4 member 2) (BND3L). | |||||
|
HSF4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.010858 (rank : 65) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULV5, Q99472, Q9ULV6 | Gene names | HSF4 | |||
|
Domain Architecture |
|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (hHSF4). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.024940 (rank : 43) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
R4RL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.002584 (rank : 76) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86UN2, Q8ND46 | Gene names | RTN4RL1, NGRH2, NGRL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4 receptor-like 1 precursor (Nogo-66 receptor homolog 2) (Nogo-66 receptor-related protein 3) (NgR3) (Nogo receptor-like 2). | |||||
|
SESN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.008919 (rank : 68) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P58004, Q96SI5 | Gene names | SESN2, SEST2 | |||
|
Domain Architecture |
|
|||||
| Description | Sestrin-2 (Hi95). | |||||
|
ZFP30_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | -0.002152 (rank : 78) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2G7, Q58EY8 | Gene names | ZFP30, KIAA0961, ZNF745 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 30 homolog (Zfp-30) (Zinc finger protein 745). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.009187 (rank : 67) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.034962 (rank : 32) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
COGA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.014938 (rank : 56) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q07092 | Gene names | COL16A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVI) chain precursor. | |||||
|
HXA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.003244 (rank : 74) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43365 | Gene names | HOXA3, HOX1E | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1E). | |||||
|
LBXCO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.009212 (rank : 66) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P84550 | Gene names | LBXCOR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladybird homeobox corepressor 1. | |||||
|
MAP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.019531 (rank : 53) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
WDR22_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.005378 (rank : 73) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JK2, O60559, Q8N3V3, Q8N3V5 | Gene names | WDR22, BCRG2, KIAA1824 | |||
|
Domain Architecture |
|
|||||
| Description | WD repeat protein 22 (Breakpoint cluster region protein 2) (BCRP2). | |||||
|
IRS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.053480 (rank : 24) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.050130 (rank : 27) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
FRS3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O43559, Q5T3D5 | Gene names | FRS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (Suc1-associated neurotrophic factor target 2) (SNT-2) (FGFR signaling adaptor SNT2). | |||||
|
FRS2_HUMAN
|
||||||
| NC score | 0.952806 (rank : 2) | θ value | 4.92331e-123 (rank : 3) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WU20, O43558, Q7LDQ6 | Gene names | FRS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 2 (FGFR substrate 2) (Suc1-associated neurotrophic factor target 1) (SNT-1) (FGFR signaling adaptor SNT). | |||||
|
FRS2_MOUSE
|
||||||
| NC score | 0.951216 (rank : 3) | θ value | 8.39787e-123 (rank : 4) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C180 | Gene names | Frs2, Frs2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 2 (FGFR substrate 2) (Suc1-associated neurotrophic factor target 1) (SNT-1) (FGFR signaling adaptor SNT) (FRS2 alpha). | |||||
|
FRS3_MOUSE
|
||||||
| NC score | 0.949636 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91WJ0 | Gene names | Frs3, Frs2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (FRS2-beta). | |||||
|
DOK1_HUMAN
|
||||||
| NC score | 0.519535 (rank : 5) | θ value | 2.13673e-09 (rank : 6) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99704, O43204, Q9UHG6 | Gene names | DOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 1 (Downstream of tyrosine kinase 1) (p62(dok)) (pp62). | |||||
|
DOK1_MOUSE
|
||||||
| NC score | 0.519437 (rank : 6) | θ value | 4.30538e-10 (rank : 5) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97465, Q9R213 | Gene names | Dok1, Dok | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 1 (Downstream of tyrosine kinase 1) (p62(dok)). | |||||
|
DOK3_MOUSE
|
||||||
| NC score | 0.516986 (rank : 7) | θ value | 1.06045e-08 (rank : 9) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QZK7 | Gene names | Dok3, Dokl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 3 (Downstream of tyrosine kinase 3) (P62(dok)-like protein) (DOK-L). | |||||
|
DOK2_HUMAN
|
||||||
| NC score | 0.508692 (rank : 8) | θ value | 2.36244e-08 (rank : 11) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60496 | Gene names | DOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 2 (Downstream of tyrosine kinase 2) (p56(dok-2)). | |||||
|
DOK3_HUMAN
|
||||||
| NC score | 0.500677 (rank : 9) | θ value | 1.38499e-08 (rank : 10) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7L591, Q8N864, Q9BQB3, Q9H666 | Gene names | DOK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 3 (Downstream of tyrosine kinase 3). | |||||
|
DOK2_MOUSE
|
||||||
| NC score | 0.480451 (rank : 10) | θ value | 1.06045e-08 (rank : 8) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O70469, O70272, Q99KL1 | Gene names | Dok2, Frip | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 2 (Downstream of tyrosine kinase 2) (p56(dok-2)) (Dok- related protein) (Dok-R) (IL-four receptor-interacting protein) (FRIP). | |||||
|
DOK6_HUMAN
|
||||||
| NC score | 0.479325 (rank : 11) | θ value | 8.11959e-09 (rank : 7) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PKX4, Q4V9S3, Q8WUZ8, Q96HI2, Q96LU2 | Gene names | DOK6, DOK5L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 6 (Downstream of tyrosine kinase 6). | |||||
|
DOK5_HUMAN
|
||||||
| NC score | 0.454897 (rank : 12) | θ value | 3.41135e-07 (rank : 14) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9P104, Q5T7Y0, Q5TE53, Q8TEW7, Q96H13, Q9BZ24, Q9NQF4, Q9Y411 | Gene names | DOK5, C20orf180 | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 5 (Downstream of tyrosine kinase 5) (IRS6) (Protein dok-5). | |||||
|
DOK5_MOUSE
|
||||||
| NC score | 0.453841 (rank : 13) | θ value | 3.41135e-07 (rank : 15) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91ZM9, Q8BRI3, Q9CSM6 | Gene names | Dok5 | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 5 (Downstream of tyrosine kinase 5) (Protein dok-5). | |||||
|
DOK4_HUMAN
|
||||||
| NC score | 0.452519 (rank : 14) | θ value | 1.17247e-07 (rank : 12) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TEW6, O75209, Q9BTP2, Q9NVV3 | Gene names | DOK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 4 (Downstream of tyrosine kinase 4). | |||||
|
DOK4_MOUSE
|
||||||
| NC score | 0.450422 (rank : 15) | θ value | 1.17247e-07 (rank : 13) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99KE3, Q3U0X3 | Gene names | Dok4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 4 (Downstream of tyrosine kinase 4). | |||||
|
DOK7_HUMAN
|
||||||
| NC score | 0.236817 (rank : 16) | θ value | 0.00869519 (rank : 19) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q18PE1, Q6P6A6, Q86XG5, Q8N2J3, Q8NBC1 | Gene names | DOK7, C4orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
DOK7_MOUSE
|
||||||
| NC score | 0.227030 (rank : 17) | θ value | 0.0252991 (rank : 21) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
PRPE_HUMAN
|
||||||
| NC score | 0.089188 (rank : 18) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P02811 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide P-E (IB-9). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.080262 (rank : 19) | θ value | 0.00509761 (rank : 18) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.068763 (rank : 20) | θ value | 0.000461057 (rank : 17) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.068745 (rank : 21) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.059965 (rank : 22) | θ value | 0.0736092 (rank : 23) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.058889 (rank : 23) | θ value | 0.0252991 (rank : 20) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
IRS1_MOUSE
|
||||||
| NC score | 0.053480 (rank : 24) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.051411 (rank : 25) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.050395 (rank : 26) | θ value | 0.0563607 (rank : 22) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.050130 (rank : 27) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
WASP_MOUSE
|
||||||
| NC score | 0.047062 (rank : 28) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.045414 (rank : 29) | θ value | 9.29e-05 (rank : 16) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
PRP5_HUMAN
|
||||||
| NC score | 0.040064 (rank : 30) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.036444 (rank : 31) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.034962 (rank : 32) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.034386 (rank : 33) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.033727 (rank : 34) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.032539 (rank : 35) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
IP3KA_MOUSE
|
||||||
| NC score | 0.032266 (rank : 36) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R071 | Gene names | Itpka | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol-trisphosphate 3-kinase A (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase A) (IP3K A) (IP3 3-kinase A). | |||||
|
SAPS1_MOUSE
|
||||||
| NC score | 0.029946 (rank : 37) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
CBL_MOUSE
|
||||||
| NC score | 0.029906 (rank : 38) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P22682, Q8CEA1 | Gene names | Cbl | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene). | |||||
|
MAP2_HUMAN
|
||||||
| NC score | 0.029887 (rank : 39) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.029684 (rank : 40) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.029594 (rank : 41) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.025837 (rank : 42) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.024940 (rank : 43) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
HNRPG_MOUSE
|
||||||
| NC score | 0.024167 (rank : 44) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35479 | Gene names | Rbmx, Hnrnpg, Hnrpg, Rbmxp1, Rbmxrt | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein G (hnRNP G) (RNA-binding motif protein, X chromosome). | |||||
|
NED4L_MOUSE
|
||||||
| NC score | 0.023744 (rank : 45) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
CLCN1_MOUSE
|
||||||
| NC score | 0.023608 (rank : 46) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64347 | Gene names | Clcn1, Clc1 | |||
|
Domain Architecture |
|
|||||
| Description | Chloride channel protein, skeletal muscle (Chloride channel protein 1) (ClC-1). | |||||
|
NED4L_HUMAN
|
||||||
| NC score | 0.023337 (rank : 47) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.022822 (rank : 48) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
HNRPG_HUMAN
|
||||||
| NC score | 0.022518 (rank : 49) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P38159, Q5JQ67, Q969R3 | Gene names | RBMX, HNRPG, RBMXP1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein G (hnRNP G) (RNA-binding motif protein, X chromosome) (Glycoprotein p43). | |||||
|
KCNH2_HUMAN
|
||||||
| NC score | 0.022513 (rank : 50) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
PER1_MOUSE
|
||||||
| NC score | 0.021478 (rank : 51) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
CSKI2_MOUSE
|
||||||
| NC score | 0.020958 (rank : 52) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
MAP2_MOUSE
|
||||||
| NC score | 0.019531 (rank : 53) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
NEDD4_HUMAN
|
||||||
| NC score | 0.016990 (rank : 54) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
CSKI2_HUMAN
|
||||||
| NC score | 0.015839 (rank : 55) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
COGA1_HUMAN
|
||||||
| NC score | 0.014938 (rank : 56) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q07092 | Gene names | COL16A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVI) chain precursor. | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.014267 (rank : 57) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
CLMN_MOUSE
|
||||||
| NC score | 0.014171 (rank : 58) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C5W0, Q91V71, Q91XT7, Q91XT8, Q91XU9 | Gene names | Clmn | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
ADA19_HUMAN
|
||||||
| NC score | 0.014096 (rank : 59) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H013, Q9BZL5, Q9UHP2 | Gene names | ADAM19, MLTNB | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 19) (Meltrin beta) (Metalloprotease and disintegrin dentritic antigen marker) (MADDAM). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.012985 (rank : 60) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
SIRT6_HUMAN
|
||||||
| NC score | 0.012829 (rank : 61) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N6T7, O75291, Q6IAF5, Q6PK99, Q8NCD2, Q9BSI5, Q9BWP3, Q9NRC7, Q9UQD1 | Gene names | SIRT6, SIR2L6 | |||
|
Domain Architecture |

|
|||||
| Description | Mono-ADP-ribosyltransferase sirtuin-6 (EC 2.4.2.31) (SIR2-like protein 6). | |||||
|
PSD4_HUMAN
|
||||||
| NC score | 0.012400 (rank : 62) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NDX1, O95621, Q4ZG34, Q6GPH8, Q8IYP4 | Gene names | PSD4, EFA6B, TIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6B) (Telomeric of interleukin-1 cluster protein). | |||||
|
ARP5_HUMAN
|
||||||
| NC score | 0.011687 (rank : 63) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H9F9, Q86WF7, Q8IUY5, Q8N724, Q9BRN0, Q9BVB7 | Gene names | ACTR5, ARP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-related protein 5 (hARP5) (Sarcoma antigen NY-SAR-16). | |||||
|
NEB2_HUMAN
|
||||||
| NC score | 0.011640 (rank : 64) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
HSF4_HUMAN
|
||||||
| NC score | 0.010858 (rank : 65) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULV5, Q99472, Q9ULV6 | Gene names | HSF4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (hHSF4). | |||||
|
LBXCO_HUMAN
|
||||||
| NC score | 0.009212 (rank : 66) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P84550 | Gene names | LBXCOR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladybird homeobox corepressor 1. | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.009187 (rank : 67) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
SESN2_HUMAN
|
||||||
| NC score | 0.008919 (rank : 68) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P58004, Q96SI5 | Gene names | SESN2, SEST2 | |||
|
Domain Architecture |

|
|||||
| Description | Sestrin-2 (Hi95). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.007945 (rank : 69) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.007476 (rank : 70) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
COIA1_MOUSE
|
||||||
| NC score | 0.007370 (rank : 71) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
B3A2_HUMAN
|
||||||
| NC score | 0.006703 (rank : 72) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04920, Q45EY5, Q969L3 | Gene names | SLC4A2, AE2, EPB3L1, HKB3, MPB3L | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 2 (Non-erythroid band 3-like protein) (AE2 anion exchanger) (Solute carrier family 4 member 2) (BND3L). | |||||
|
WDR22_HUMAN
|
||||||
| NC score | 0.005378 (rank : 73) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JK2, O60559, Q8N3V3, Q8N3V5 | Gene names | WDR22, BCRG2, KIAA1824 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 22 (Breakpoint cluster region protein 2) (BCRP2). | |||||
|
HXA3_HUMAN
|
||||||
| NC score | 0.003244 (rank : 74) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43365 | Gene names | HOXA3, HOX1E | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1E). | |||||
|
ACK1_HUMAN
|
||||||
| NC score | 0.002660 (rank : 75) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1140 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q07912, Q8N6U7, Q96H59 | Gene names | TNK2, ACK1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Tyrosine kinase non- receptor protein 2). | |||||
|
R4RL1_HUMAN
|
||||||
| NC score | 0.002584 (rank : 76) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86UN2, Q8ND46 | Gene names | RTN4RL1, NGRH2, NGRL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4 receptor-like 1 precursor (Nogo-66 receptor homolog 2) (Nogo-66 receptor-related protein 3) (NgR3) (Nogo receptor-like 2). | |||||
|
TAOK2_MOUSE
|
||||||
| NC score | 0.001406 (rank : 77) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1373 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZQ29, Q7TSS8 | Gene names | Taok2, Kiaa0881 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2). | |||||
|
ZFP30_HUMAN
|
||||||
| NC score | -0.002152 (rank : 78) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2G7, Q58EY8 | Gene names | ZFP30, KIAA0961, ZNF745 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 30 homolog (Zfp-30) (Zinc finger protein 745). | |||||