Please be patient as the page loads
|
NMDE4_HUMAN
|
||||||
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NMDE1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.984288 (rank : 8) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q12879, O00669 | Gene names | GRIN2A | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A) (hNR2A). | |||||
|
NMDE1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.984920 (rank : 5) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P35436 | Gene names | Grin2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A). | |||||
|
NMDE2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.984420 (rank : 6) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13224, Q12919, Q13220, Q13225, Q9UM56 | Gene names | GRIN2B | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B) (N-methyl-D-aspartate receptor subunit 3) (NR3) (hNR3). | |||||
|
NMDE2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.984362 (rank : 7) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q01097, Q9DCB2 | Gene names | Grin2b | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B). | |||||
|
NMDE3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.988702 (rank : 4) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.989017 (rank : 3) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q01098 | Gene names | Grin2c | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE4_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 133 | |
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
|
NMDE4_MOUSE
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.995669 (rank : 2) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q03391 | Gene names | Grin2d | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D). | |||||
|
NMD3A_HUMAN
|
||||||
| θ value | 1.11217e-74 (rank : 9) | NC score | 0.935385 (rank : 9) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8TCU5, Q8TF29, Q8WXI6 | Gene names | GRIN3A, KIAA1973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3A precursor (N-methyl-D-aspartate receptor subtype NR3A) (NMDAR-L). | |||||
|
NMDZ1_MOUSE
|
||||||
| θ value | 1.27248e-70 (rank : 10) | NC score | 0.910076 (rank : 13) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35438, Q8CFS4 | Gene names | Grin1, Glurz1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMDZ1_HUMAN
|
||||||
| θ value | 3.70236e-70 (rank : 11) | NC score | 0.910436 (rank : 12) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q05586, P35437, Q12867, Q12868 | Gene names | GRIN1, NMDAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMD3B_MOUSE
|
||||||
| θ value | 1.5555e-68 (rank : 12) | NC score | 0.927980 (rank : 10) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91ZU9, Q8VHV0 | Gene names | Grin3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subunit NR3B) (NR3B) (NMDAR3B) (NMDA receptor NR4) (Nr4). | |||||
|
NMD3B_HUMAN
|
||||||
| θ value | 1.11475e-66 (rank : 13) | NC score | 0.927043 (rank : 11) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O60391, Q5EAK7, Q7RTW9 | Gene names | GRIN3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subtype NR3B) (NR3B) (NMDAR3B). | |||||
|
GRIK3_HUMAN
|
||||||
| θ value | 2.58187e-47 (rank : 14) | NC score | 0.847194 (rank : 17) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13003, Q13004, Q16136 | Gene names | GRIK3, GLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 3 precursor (Glutamate receptor 7) (GluR-7) (GluR7) (Excitatory amino acid receptor 5) (EAA5). | |||||
|
GRIK5_MOUSE
|
||||||
| θ value | 4.86918e-46 (rank : 15) | NC score | 0.845258 (rank : 23) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61626 | Gene names | Grik5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Glutamate receptor gamma-2) (GluR gamma-2). | |||||
|
GRIK5_HUMAN
|
||||||
| θ value | 1.85029e-45 (rank : 16) | NC score | 0.845506 (rank : 22) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q16478 | Gene names | GRIK5, GRIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Excitatory amino acid receptor 2) (EAA2). | |||||
|
GRIK1_HUMAN
|
||||||
| θ value | 9.18288e-45 (rank : 17) | NC score | 0.846083 (rank : 20) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P39086, Q13001, Q86SU9 | Gene names | GRIK1, GLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5) (Excitatory amino acid receptor 3) (EAA3). | |||||
|
GRIK4_HUMAN
|
||||||
| θ value | 1.01529e-43 (rank : 18) | NC score | 0.846411 (rank : 18) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q16099 | Gene names | GRIK4, GRIK | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor (Glutamate receptor KA-1) (KA1) (Excitatory amino acid receptor 1) (EAA1). | |||||
|
GRIK4_MOUSE
|
||||||
| θ value | 1.01529e-43 (rank : 19) | NC score | 0.846103 (rank : 19) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BMF5 | Gene names | Grik4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor. | |||||
|
GRIA1_HUMAN
|
||||||
| θ value | 1.73182e-43 (rank : 20) | NC score | 0.838667 (rank : 26) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P42261 | Gene names | GRIA1, GLUH1, GLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIA1_MOUSE
|
||||||
| θ value | 1.73182e-43 (rank : 21) | NC score | 0.839934 (rank : 25) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P23818 | Gene names | Gria1, Glur1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIK2_HUMAN
|
||||||
| θ value | 1.73182e-43 (rank : 22) | NC score | 0.844427 (rank : 24) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13002 | Gene names | GRIK2, GLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 2 precursor (Glutamate receptor 6) (GluR-6) (GluR6) (Excitatory amino acid receptor 4) (EAA4). | |||||
|
GRIA4_HUMAN
|
||||||
| θ value | 3.85808e-43 (rank : 23) | NC score | 0.835953 (rank : 27) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48058 | Gene names | GRIA4, GLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIA2_HUMAN
|
||||||
| θ value | 1.46607e-42 (rank : 24) | NC score | 0.834883 (rank : 28) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P42262, Q96FP6 | Gene names | GRIA2, GLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIA2_MOUSE
|
||||||
| θ value | 4.2656e-42 (rank : 25) | NC score | 0.834205 (rank : 29) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P23819, Q61604, Q61605, Q8BG69, Q8BXU3, Q9D6D3 | Gene names | Gria2, Glur2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRID2_MOUSE
|
||||||
| θ value | 7.27602e-42 (rank : 26) | NC score | 0.851030 (rank : 14) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61625 | Gene names | Grid2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRIA3_MOUSE
|
||||||
| θ value | 1.62093e-41 (rank : 27) | NC score | 0.833858 (rank : 30) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z2W9, Q5DTJ0 | Gene names | Gria3, Glur3, Kiaa4184 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRIA4_MOUSE
|
||||||
| θ value | 2.11701e-41 (rank : 28) | NC score | 0.833768 (rank : 31) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2W8 | Gene names | Gria4, Glur4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRID1_MOUSE
|
||||||
| θ value | 2.11701e-41 (rank : 29) | NC score | 0.847445 (rank : 16) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61627 | Gene names | Grid1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRID2_HUMAN
|
||||||
| θ value | 2.11701e-41 (rank : 30) | NC score | 0.849931 (rank : 15) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43424 | Gene names | GRID2, GLURD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRIA3_HUMAN
|
||||||
| θ value | 8.04462e-41 (rank : 31) | NC score | 0.833659 (rank : 32) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P42263, Q4VXD5, Q4VXD6, Q9HDA0, Q9HDA1, Q9HDA2 | Gene names | GRIA3, GLUR3, GLURC | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRID1_HUMAN
|
||||||
| θ value | 2.34062e-40 (rank : 32) | NC score | 0.845772 (rank : 21) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULK0, Q8IXT3 | Gene names | GRID1, KIAA1220 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRIK1_MOUSE
|
||||||
| θ value | 1.12514e-34 (rank : 33) | NC score | 0.828064 (rank : 33) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q60934 | Gene names | Grik1, Glur5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 34) | NC score | 0.036861 (rank : 39) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 35) | NC score | 0.014263 (rank : 73) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 36) | NC score | 0.039400 (rank : 38) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 37) | NC score | 0.021064 (rank : 51) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 38) | NC score | 0.028506 (rank : 47) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.125558 (rank : 39) | NC score | 0.015975 (rank : 68) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 40) | NC score | 0.005557 (rank : 110) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
FOXE2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 41) | NC score | 0.014135 (rank : 74) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99526 | Gene names | FOXE2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E2 (HNF-3/fork head-like protein 5) (HFKL5) (HFKH4). | |||||
|
GABR1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 42) | NC score | 0.072748 (rank : 35) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 43) | NC score | 0.072700 (rank : 36) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 44) | NC score | 0.035235 (rank : 42) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
ERB1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 45) | NC score | 0.133162 (rank : 34) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60509 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-R(b)_3p24.3 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-R(b) Env protein) [Includes: Surface protein domain (SU); Transmembrane protein domain (TM)]. | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 0.21417 (rank : 46) | NC score | 0.018254 (rank : 58) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 0.279714 (rank : 47) | NC score | 0.036448 (rank : 40) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.019729 (rank : 53) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 49) | NC score | 0.031000 (rank : 44) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
TS1R3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | 0.030369 (rank : 45) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7RTX0, Q5TA49, Q8NGW9 | Gene names | TAS1R3, T1R3, TR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 3 precursor (Sweet taste receptor T1R3). | |||||
|
ANPRB_HUMAN
|
||||||
| θ value | 0.62314 (rank : 51) | NC score | 0.005693 (rank : 109) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20594, O60871, Q9UQ50 | Gene names | NPR2, ANPRB | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
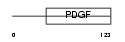
VEGFB_HUMAN
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.028966 (rank : 46) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49765, Q16528 | Gene names | VEGFB, VRF | |||
|
Domain Architecture |
|
|||||
| Description | Vascular endothelial growth factor B precursor (VEGF-B) (VEGF-related factor) (VRF). | |||||
|
ELL_HUMAN
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.021919 (rank : 49) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55199 | Gene names | ELL, C19orf17 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL (Eleven-nineteen lysine-rich leukemia protein). | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.018148 (rank : 59) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.028155 (rank : 48) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
EGFL9_MOUSE
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.003230 (rank : 116) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K1E3, Q9QYP3 | Gene names | Egfl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9) (Endothelial cell-specific protein S-1). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.018700 (rank : 55) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
ANPRB_MOUSE
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.005263 (rank : 112) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6VVW5, Q6VVW3, Q6VVW4, Q8CGA9 | Gene names | Npr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
CCDC9_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.016742 (rank : 66) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
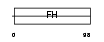
FOXD4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.012065 (rank : 80) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12950, Q5VVK1, Q8WXT6 | Gene names | FOXD4, FKHL9, FOXD4A, FREAC5 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein D4 (Forkhead-related protein FKHL9) (Forkhead- related transcription factor 5) (FREAC-5) (Myeloid factor-alpha). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.013721 (rank : 75) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.008666 (rank : 97) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.021849 (rank : 50) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
GAK5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.016796 (rank : 65) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
TS1R1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.050267 (rank : 37) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99PG6, Q6NS58, Q923J9, Q925I5, Q99PG5 | Gene names | Tas1r1, Gpr70, T1r1, Tr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70). | |||||
|
ZO3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.008076 (rank : 101) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95049 | Gene names | TJP3, ZO3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
ATX2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.019579 (rank : 54) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
CELR2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.000071 (rank : 125) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
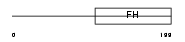
FX4L1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.011921 (rank : 82) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NU39 | Gene names | FOXD4L1 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein D4-like 1. | |||||
|
IQEC3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.008562 (rank : 100) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.009269 (rank : 95) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MEGF8_MOUSE
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.008579 (rank : 99) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.012944 (rank : 77) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.017356 (rank : 62) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
RFIP5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.011977 (rank : 81) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R361 | Gene names | Rab11fip5, D6Ertd32e, Rip11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 5 (Rab11-FIP5) (Rab11-interacting protein Rip11). | |||||
|
CO4A2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.006993 (rank : 105) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
MYO9B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.002704 (rank : 119) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
SRGEF_MOUSE
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.010188 (rank : 88) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80YD6, Q9QXB7 | Gene names | Sergef, Delgef, Gnefr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
ATS13_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.006538 (rank : 108) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
CK024_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.035544 (rank : 41) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
DOK7_MOUSE
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.010605 (rank : 87) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
EGR4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | -0.003363 (rank : 132) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
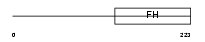
FOXD3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.009331 (rank : 92) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61060 | Gene names | Foxd3, Hfh2 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein D3 (HNF3/FH transcription factor genesis) (Hepatocyte nuclear factor 3 forkhead homolog 2) (HFH-2). | |||||
|
GAK11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.015421 (rank : 71) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.015456 (rank : 69) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.015412 (rank : 72) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.015426 (rank : 70) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GGA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.005073 (rank : 113) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
HRBL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.008663 (rank : 98) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.012611 (rank : 78) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PCDA9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.001578 (rank : 121) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y5H5, O15053 | Gene names | PCDHA9, KIAA0345 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 9 precursor (PCDH-alpha9). | |||||
|
ARFP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.007444 (rank : 103) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P53367, Q9Y2X6 | Gene names | ARFIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Arfaptin-1 (ADP-ribosylation factor-interacting protein 1). | |||||
|
BIM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.009821 (rank : 90) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43521, O43522 | Gene names | BCL2L11, BIM | |||
|
Domain Architecture |

|
|||||
| Description | Bcl-2-like protein 11 (Bcl2-interacting mediator of cell death). | |||||
|
CRSP7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.010921 (rank : 84) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | -0.002176 (rank : 129) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
FOXN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.009107 (rank : 96) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O15353, O15352 | Gene names | FOXN1, WHN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude). | |||||
|
FOXN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.017880 (rank : 60) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.033110 (rank : 43) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
LHX2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.003077 (rank : 118) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P50458, O95860, Q8N1Z3 | Gene names | LHX2, LH2 | |||
|
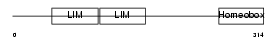
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx2 (Homeobox protein LH-2). | |||||
|
LHX2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.003092 (rank : 117) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0S2 | Gene names | Lhx2 | |||
|
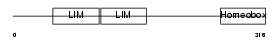
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx2. | |||||
|
PEX14_MOUSE
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.010137 (rank : 89) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0A0 | Gene names | Pex14 | |||
|
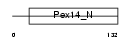
Domain Architecture |
|
|||||
| Description | Peroxisomal membrane protein PEX14 (Peroxin-14) (Peroxisomal membrane anchor protein PEX14) (PTS1 receptor docking protein). | |||||
|
PGBM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.000101 (rank : 124) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
PGBM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.000945 (rank : 122) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | -0.000764 (rank : 127) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.017796 (rank : 61) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CBP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.017123 (rank : 64) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CO5A3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.006771 (rank : 107) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P25940, Q9NZQ6 | Gene names | COL5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(V) chain precursor. | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.017198 (rank : 63) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
GAK10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.013030 (rank : 76) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.011459 (rank : 83) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
MEGF8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.007135 (rank : 104) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.019934 (rank : 52) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.016312 (rank : 67) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
ANDR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.000047 (rank : 126) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P19091 | Gene names | Ar, Nr3c4 | |||
|
Domain Architecture |

|
|||||
| Description | Androgen receptor (Dihydrotestosterone receptor). | |||||
|
ARI5B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.004857 (rank : 114) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
CIAS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.002586 (rank : 120) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96P20, O75434, Q59H68, Q5JQS8, Q5JQS9, Q6TG35, Q8TCW0, Q8TEU9, Q8WXH9 | Gene names | CIAS1, NALP3, PYPAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein (Cryopyrin) (NACHT-, LRR- and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.018424 (rank : 57) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
DRD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | -0.002978 (rank : 131) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51436, O35838 | Gene names | Drd4 | |||
|
Domain Architecture |

|
|||||
| Description | D(4) dopamine receptor (Dopamine D4 receptor) (D(2C) dopamine receptor). | |||||
|
FX4L4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.007449 (rank : 102) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WXT5 | Gene names | FOXD4L4, FOXD4B | |||
|
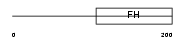
Domain Architecture |
|
|||||
| Description | Forkhead box protein D4-like 4 (Forkhead box protein D4B) (Myeloid factor-gamma). | |||||
|
GAK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | -0.002315 (rank : 130) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
GP152_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.005276 (rank : 111) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
HXA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | -0.001462 (rank : 128) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31245, Q149U3, Q920T8 | Gene names | Hoxa2, Hox-1.11, Hoxa-2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A2 (Hox-1.11) (Hox1.11). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.010639 (rank : 86) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
KTNA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.000628 (rank : 123) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75449, Q5TFA8, Q5TFA9, Q86VN2, Q9NU52 | Gene names | KATNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin). | |||||
|
MGR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.018435 (rank : 56) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14416, Q52MC6, Q9H3N6 | Gene names | GRM2, GPRC1B, MGLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 2 precursor (mGluR2). | |||||
|
RS3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.009308 (rank : 94) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23396, Q498B5, Q8NI95 | Gene names | RPS3 | |||
|
Domain Architecture |
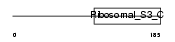
|
|||||
| Description | 40S ribosomal protein S3. | |||||
|
RS3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.009308 (rank : 93) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62908, P17073, P47933 | Gene names | Rps3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 40S ribosomal protein S3. | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.009621 (rank : 91) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
SLY_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.003336 (rank : 115) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K352, Q3TBX0, Q9DBV2 | Gene names | Sly | |||
|
Domain Architecture |
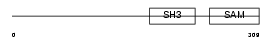
|
|||||
| Description | SH3 protein expressed in lymphocytes. | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.010787 (rank : 85) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
TNR6A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.006920 (rank : 106) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
VEGFB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.012583 (rank : 79) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49766, Q3UG04, Q5D0B1, Q64290 | Gene names | Vegfb, Vrf | |||
|
Domain Architecture |
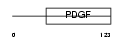
|
|||||
| Description | Vascular endothelial growth factor B precursor (VEGF-B) (VEGF-related factor) (VRF). | |||||
|
ZN167_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | -0.005194 (rank : 133) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P0L1, Q6WL09, Q96FQ2 | Gene names | ZNF167, ZNF448, ZNF64 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 167 (Zinc finger protein 64) (Zinc finger protein 448) (ZFP). | |||||
|
NMDE4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 133 | |
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
|
NMDE4_MOUSE
|
||||||
| NC score | 0.995669 (rank : 2) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q03391 | Gene names | Grin2d | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D). | |||||
|
NMDE3_MOUSE
|
||||||
| NC score | 0.989017 (rank : 3) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q01098 | Gene names | Grin2c | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE3_HUMAN
|
||||||
| NC score | 0.988702 (rank : 4) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE1_MOUSE
|
||||||
| NC score | 0.984920 (rank : 5) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P35436 | Gene names | Grin2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A). | |||||
|
NMDE2_HUMAN
|
||||||
| NC score | 0.984420 (rank : 6) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13224, Q12919, Q13220, Q13225, Q9UM56 | Gene names | GRIN2B | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B) (N-methyl-D-aspartate receptor subunit 3) (NR3) (hNR3). | |||||
|
NMDE2_MOUSE
|
||||||
| NC score | 0.984362 (rank : 7) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q01097, Q9DCB2 | Gene names | Grin2b | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B). | |||||
|
NMDE1_HUMAN
|
||||||
| NC score | 0.984288 (rank : 8) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q12879, O00669 | Gene names | GRIN2A | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A) (hNR2A). | |||||
|
NMD3A_HUMAN
|
||||||
| NC score | 0.935385 (rank : 9) | θ value | 1.11217e-74 (rank : 9) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8TCU5, Q8TF29, Q8WXI6 | Gene names | GRIN3A, KIAA1973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3A precursor (N-methyl-D-aspartate receptor subtype NR3A) (NMDAR-L). | |||||
|
NMD3B_MOUSE
|
||||||
| NC score | 0.927980 (rank : 10) | θ value | 1.5555e-68 (rank : 12) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91ZU9, Q8VHV0 | Gene names | Grin3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subunit NR3B) (NR3B) (NMDAR3B) (NMDA receptor NR4) (Nr4). | |||||
|
NMD3B_HUMAN
|
||||||
| NC score | 0.927043 (rank : 11) | θ value | 1.11475e-66 (rank : 13) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O60391, Q5EAK7, Q7RTW9 | Gene names | GRIN3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subtype NR3B) (NR3B) (NMDAR3B). | |||||
|
NMDZ1_HUMAN
|
||||||
| NC score | 0.910436 (rank : 12) | θ value | 3.70236e-70 (rank : 11) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q05586, P35437, Q12867, Q12868 | Gene names | GRIN1, NMDAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMDZ1_MOUSE
|
||||||
| NC score | 0.910076 (rank : 13) | θ value | 1.27248e-70 (rank : 10) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35438, Q8CFS4 | Gene names | Grin1, Glurz1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
GRID2_MOUSE
|
||||||
| NC score | 0.851030 (rank : 14) | θ value | 7.27602e-42 (rank : 26) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61625 | Gene names | Grid2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRID2_HUMAN
|
||||||
| NC score | 0.849931 (rank : 15) | θ value | 2.11701e-41 (rank : 30) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43424 | Gene names | GRID2, GLURD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRID1_MOUSE
|
||||||
| NC score | 0.847445 (rank : 16) | θ value | 2.11701e-41 (rank : 29) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61627 | Gene names | Grid1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRIK3_HUMAN
|
||||||
| NC score | 0.847194 (rank : 17) | θ value | 2.58187e-47 (rank : 14) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13003, Q13004, Q16136 | Gene names | GRIK3, GLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 3 precursor (Glutamate receptor 7) (GluR-7) (GluR7) (Excitatory amino acid receptor 5) (EAA5). | |||||
|
GRIK4_HUMAN
|
||||||
| NC score | 0.846411 (rank : 18) | θ value | 1.01529e-43 (rank : 18) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q16099 | Gene names | GRIK4, GRIK | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor (Glutamate receptor KA-1) (KA1) (Excitatory amino acid receptor 1) (EAA1). | |||||
|
GRIK4_MOUSE
|
||||||
| NC score | 0.846103 (rank : 19) | θ value | 1.01529e-43 (rank : 19) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BMF5 | Gene names | Grik4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor. | |||||
|
GRIK1_HUMAN
|
||||||
| NC score | 0.846083 (rank : 20) | θ value | 9.18288e-45 (rank : 17) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P39086, Q13001, Q86SU9 | Gene names | GRIK1, GLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5) (Excitatory amino acid receptor 3) (EAA3). | |||||
|
GRID1_HUMAN
|
||||||
| NC score | 0.845772 (rank : 21) | θ value | 2.34062e-40 (rank : 32) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULK0, Q8IXT3 | Gene names | GRID1, KIAA1220 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRIK5_HUMAN
|
||||||
| NC score | 0.845506 (rank : 22) | θ value | 1.85029e-45 (rank : 16) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q16478 | Gene names | GRIK5, GRIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Excitatory amino acid receptor 2) (EAA2). | |||||
|
GRIK5_MOUSE
|
||||||
| NC score | 0.845258 (rank : 23) | θ value | 4.86918e-46 (rank : 15) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61626 | Gene names | Grik5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Glutamate receptor gamma-2) (GluR gamma-2). | |||||
|
GRIK2_HUMAN
|
||||||
| NC score | 0.844427 (rank : 24) | θ value | 1.73182e-43 (rank : 22) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13002 | Gene names | GRIK2, GLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 2 precursor (Glutamate receptor 6) (GluR-6) (GluR6) (Excitatory amino acid receptor 4) (EAA4). | |||||
|
GRIA1_MOUSE
|
||||||
| NC score | 0.839934 (rank : 25) | θ value | 1.73182e-43 (rank : 21) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P23818 | Gene names | Gria1, Glur1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIA1_HUMAN
|
||||||
| NC score | 0.838667 (rank : 26) | θ value | 1.73182e-43 (rank : 20) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P42261 | Gene names | GRIA1, GLUH1, GLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIA4_HUMAN
|
||||||
| NC score | 0.835953 (rank : 27) | θ value | 3.85808e-43 (rank : 23) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48058 | Gene names | GRIA4, GLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIA2_HUMAN
|
||||||
| NC score | 0.834883 (rank : 28) | θ value | 1.46607e-42 (rank : 24) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P42262, Q96FP6 | Gene names | GRIA2, GLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIA2_MOUSE
|
||||||
| NC score | 0.834205 (rank : 29) | θ value | 4.2656e-42 (rank : 25) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P23819, Q61604, Q61605, Q8BG69, Q8BXU3, Q9D6D3 | Gene names | Gria2, Glur2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIA3_MOUSE
|
||||||
| NC score | 0.833858 (rank : 30) | θ value | 1.62093e-41 (rank : 27) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z2W9, Q5DTJ0 | Gene names | Gria3, Glur3, Kiaa4184 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRIA4_MOUSE
|
||||||
| NC score | 0.833768 (rank : 31) | θ value | 2.11701e-41 (rank : 28) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2W8 | Gene names | Gria4, Glur4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIA3_HUMAN
|
||||||
| NC score | 0.833659 (rank : 32) | θ value | 8.04462e-41 (rank : 31) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P42263, Q4VXD5, Q4VXD6, Q9HDA0, Q9HDA1, Q9HDA2 | Gene names | GRIA3, GLUR3, GLURC | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRIK1_MOUSE
|
||||||
| NC score | 0.828064 (rank : 33) | θ value | 1.12514e-34 (rank : 33) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q60934 | Gene names | Grik1, Glur5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5). | |||||
|
ERB1_HUMAN
|
||||||
| NC score | 0.133162 (rank : 34) | θ value | 0.21417 (rank : 45) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60509 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-R(b)_3p24.3 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-R(b) Env protein) [Includes: Surface protein domain (SU); Transmembrane protein domain (TM)]. | |||||
|
GABR1_HUMAN
|
||||||
| NC score | 0.072748 (rank : 35) | θ value | 0.163984 (rank : 42) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_MOUSE
|
||||||
| NC score | 0.072700 (rank : 36) | θ value | 0.163984 (rank : 43) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
TS1R1_MOUSE
|
||||||
| NC score | 0.050267 (rank : 37) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99PG6, Q6NS58, Q923J9, Q925I5, Q99PG5 | Gene names | Tas1r1, Gpr70, T1r1, Tr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.039400 (rank : 38) | θ value | 0.00298849 (rank : 36) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.036861 (rank : 39) | θ value | 3.19293e-05 (rank : 34) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.036448 (rank : 40) | θ value | 0.279714 (rank : 47) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.035544 (rank : 41) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.035235 (rank : 42) | θ value | 0.163984 (rank : 44) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.033110 (rank : 43) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.031000 (rank : 44) | θ value | 0.47712 (rank : 49) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
TS1R3_HUMAN
|
||||||
| NC score | 0.030369 (rank : 45) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7RTX0, Q5TA49, Q8NGW9 | Gene names | TAS1R3, T1R3, TR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 3 precursor (Sweet taste receptor T1R3). | |||||
|
VEGFB_HUMAN
|
||||||
| NC score | 0.028966 (rank : 46) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49765, Q16528 | Gene names | VEGFB, VRF | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor B precursor (VEGF-B) (VEGF-related factor) (VRF). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.028506 (rank : 47) | θ value | 0.0736092 (rank : 38) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.028155 (rank : 48) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
ELL_HUMAN
|
||||||
| NC score | 0.021919 (rank : 49) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55199 | Gene names | ELL, C19orf17 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL (Eleven-nineteen lysine-rich leukemia protein). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.021849 (rank : 50) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.021064 (rank : 51) | θ value | 0.0431538 (rank : 37) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.019934 (rank : 52) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.019729 (rank : 53) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
ATX2_HUMAN
|
||||||
| NC score | 0.019579 (rank : 54) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.018700 (rank : 55) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
MGR2_HUMAN
|
||||||
| NC score | 0.018435 (rank : 56) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14416, Q52MC6, Q9H3N6 | Gene names | GRM2, GPRC1B, MGLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 2 precursor (mGluR2). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.018424 (rank : 57) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.018254 (rank : 58) | θ value | 0.21417 (rank : 46) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.018148 (rank : 59) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
FOXN1_MOUSE
|
||||||
| NC score | 0.017880 (rank : 60) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.017796 (rank : 61) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.017356 (rank : 62) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.017198 (rank : 63) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.017123 (rank : 64) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
GAK5_HUMAN
|
||||||
| NC score | 0.016796 (rank : 65) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
CCDC9_MOUSE
|
||||||
| NC score | 0.016742 (rank : 66) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.016312 (rank : 67) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.015975 (rank : 68) | θ value | 0.125558 (rank : 39) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
GAK12_HUMAN
|
||||||
| NC score | 0.015456 (rank : 69) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| NC score | 0.015426 (rank : 70) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK11_HUMAN
|
||||||
| NC score | 0.015421 (rank : 71) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.015412 (rank : 72) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.014263 (rank : 73) | θ value | 0.00175202 (rank : 35) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
FOXE2_HUMAN
|
||||||
| NC score | 0.014135 (rank : 74) | θ value | 0.163984 (rank : 41) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99526 | Gene names | FOXE2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E2 (HNF-3/fork head-like protein 5) (HFKL5) (HFKH4). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.013721 (rank : 75) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
GAK10_HUMAN
|
||||||
| NC score | 0.013030 (rank : 76) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.012944 (rank : 77) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.012611 (rank : 78) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
VEGFB_MOUSE
|
||||||
| NC score | 0.012583 (rank : 79) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49766, Q3UG04, Q5D0B1, Q64290 | Gene names | Vegfb, Vrf | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor B precursor (VEGF-B) (VEGF-related factor) (VRF). | |||||
|
FOXD4_HUMAN
|
||||||
| NC score | 0.012065 (rank : 80) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12950, Q5VVK1, Q8WXT6 | Gene names | FOXD4, FKHL9, FOXD4A, FREAC5 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D4 (Forkhead-related protein FKHL9) (Forkhead- related transcription factor 5) (FREAC-5) (Myeloid factor-alpha). | |||||
|
RFIP5_MOUSE
|
||||||
| NC score | 0.011977 (rank : 81) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R361 | Gene names | Rab11fip5, D6Ertd32e, Rip11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 5 (Rab11-FIP5) (Rab11-interacting protein Rip11). | |||||
|
FX4L1_HUMAN
|
||||||
| NC score | 0.011921 (rank : 82) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NU39 | Gene names | FOXD4L1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D4-like 1. | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.011459 (rank : 83) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
CRSP7_HUMAN
|
||||||
| NC score | 0.010921 (rank : 84) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.010787 (rank : 85) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.010639 (rank : 86) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
DOK7_MOUSE
|
||||||
| NC score | 0.010605 (rank : 87) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
SRGEF_MOUSE
|
||||||
| NC score | 0.010188 (rank : 88) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80YD6, Q9QXB7 | Gene names | Sergef, Delgef, Gnefr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
PEX14_MOUSE
|
||||||
| NC score | 0.010137 (rank : 89) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0A0 | Gene names | Pex14 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal membrane protein PEX14 (Peroxin-14) (Peroxisomal membrane anchor protein PEX14) (PTS1 receptor docking protein). | |||||
|
BIM_HUMAN
|
||||||
| NC score | 0.009821 (rank : 90) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43521, O43522 | Gene names | BCL2L11, BIM | |||
|
Domain Architecture |

|
|||||
| Description | Bcl-2-like protein 11 (Bcl2-interacting mediator of cell death). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.009621 (rank : 91) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
FOXD3_MOUSE
|
||||||
| NC score | 0.009331 (rank : 92) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61060 | Gene names | Foxd3, Hfh2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D3 (HNF3/FH transcription factor genesis) (Hepatocyte nuclear factor 3 forkhead homolog 2) (HFH-2). | |||||
|
RS3_MOUSE
|
||||||
| NC score | 0.009308 (rank : 93) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62908, P17073, P47933 | Gene names | Rps3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 40S ribosomal protein S3. | |||||
|
RS3_HUMAN
|
||||||
| NC score | 0.009308 (rank : 94) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23396, Q498B5, Q8NI95 | Gene names | RPS3 | |||
|
Domain Architecture |

|
|||||
| Description | 40S ribosomal protein S3. | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.009269 (rank : 95) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
FOXN1_HUMAN
|
||||||
| NC score | 0.009107 (rank : 96) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O15353, O15352 | Gene names | FOXN1, WHN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude). | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.008666 (rank : 97) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
HRBL_HUMAN
|
||||||
| NC score | 0.008663 (rank : 98) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
MEGF8_MOUSE
|
||||||
| NC score | 0.008579 (rank : 99) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
IQEC3_MOUSE
|
||||||
| NC score | 0.008562 (rank : 100) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
ZO3_HUMAN
|
||||||
| NC score | 0.008076 (rank : 101) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95049 | Gene names | TJP3, ZO3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
FX4L4_HUMAN
|
||||||
| NC score | 0.007449 (rank : 102) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WXT5 | Gene names | FOXD4L4, FOXD4B | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D4-like 4 (Forkhead box protein D4B) (Myeloid factor-gamma). | |||||
|
ARFP1_HUMAN
|
||||||
| NC score | 0.007444 (rank : 103) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P53367, Q9Y2X6 | Gene names | ARFIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Arfaptin-1 (ADP-ribosylation factor-interacting protein 1). | |||||
|
MEGF8_HUMAN
|
||||||
| NC score | 0.007135 (rank : 104) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
CO4A2_MOUSE
|
||||||
| NC score | 0.006993 (rank : 105) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
TNR6A_HUMAN
|
||||||
| NC score | 0.006920 (rank : 106) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
CO5A3_HUMAN
|
||||||
| NC score | 0.006771 (rank : 107) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P25940, Q9NZQ6 | Gene names | COL5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(V) chain precursor. | |||||
|
ATS13_HUMAN
|
||||||
| NC score | 0.006538 (rank : 108) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
ANPRB_HUMAN
|
||||||
| NC score | 0.005693 (rank : 109) | θ value | 0.62314 (rank : 51) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20594, O60871, Q9UQ50 | Gene names | NPR2, ANPRB | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.005557 (rank : 110) | θ value | 0.125558 (rank : 40) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
GP152_HUMAN
|
||||||
| NC score | 0.005276 (rank : 111) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
ANPRB_MOUSE
|
||||||
| NC score | 0.005263 (rank : 112) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6VVW5, Q6VVW3, Q6VVW4, Q8CGA9 | Gene names | Npr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
GGA1_HUMAN
|
||||||
| NC score | 0.005073 (rank : 113) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
ARI5B_HUMAN
|
||||||
| NC score | 0.004857 (rank : 114) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
SLY_MOUSE
|
||||||
| NC score | 0.003336 (rank : 115) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K352, Q3TBX0, Q9DBV2 | Gene names | Sly | |||
|
Domain Architecture |

|
|||||
| Description | SH3 protein expressed in lymphocytes. | |||||
|
EGFL9_MOUSE
|
||||||
| NC score | 0.003230 (rank : 116) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K1E3, Q9QYP3 | Gene names | Egfl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9) (Endothelial cell-specific protein S-1). | |||||
|
LHX2_MOUSE
|
||||||
| NC score | 0.003092 (rank : 117) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0S2 | Gene names | Lhx2 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx2. | |||||
|
LHX2_HUMAN
|
||||||
| NC score | 0.003077 (rank : 118) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P50458, O95860, Q8N1Z3 | Gene names | LHX2, LH2 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx2 (Homeobox protein LH-2). | |||||
|
MYO9B_MOUSE
|
||||||
| NC score | 0.002704 (rank : 119) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
CIAS1_HUMAN
|
||||||
| NC score | 0.002586 (rank : 120) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96P20, O75434, Q59H68, Q5JQS8, Q5JQS9, Q6TG35, Q8TCW0, Q8TEU9, Q8WXH9 | Gene names | CIAS1, NALP3, PYPAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein (Cryopyrin) (NACHT-, LRR- and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like). | |||||
|
PCDA9_HUMAN
|
||||||
| NC score | 0.001578 (rank : 121) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y5H5, O15053 | Gene names | PCDHA9, KIAA0345 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 9 precursor (PCDH-alpha9). | |||||
|
PGBM_MOUSE
|
||||||
| NC score | 0.000945 (rank : 122) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
KTNA1_HUMAN
|
||||||
| NC score | 0.000628 (rank : 123) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75449, Q5TFA8, Q5TFA9, Q86VN2, Q9NU52 | Gene names | KATNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin). | |||||
|
PGBM_HUMAN
|
||||||
| NC score | 0.000101 (rank : 124) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
CELR2_MOUSE
|
||||||
| NC score | 0.000071 (rank : 125) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
ANDR_MOUSE
|
||||||
| NC score | 0.000047 (rank : 126) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P19091 | Gene names | Ar, Nr3c4 | |||
|
Domain Architecture |

|
|||||
| Description | Androgen receptor (Dihydrotestosterone receptor). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | -0.000764 (rank : 127) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
HXA2_MOUSE
|
||||||
| NC score | -0.001462 (rank : 128) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31245, Q149U3, Q920T8 | Gene names | Hoxa2, Hox-1.11, Hoxa-2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A2 (Hox-1.11) (Hox1.11). | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | -0.002176 (rank : 129) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
GAK_MOUSE
|
||||||
| NC score | -0.002315 (rank : 130) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
DRD4_MOUSE
|
||||||
| NC score | -0.002978 (rank : 131) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51436, O35838 | Gene names | Drd4 | |||
|
Domain Architecture |

|
|||||
| Description | D(4) dopamine receptor (Dopamine D4 receptor) (D(2C) dopamine receptor). | |||||
|
EGR4_HUMAN
|
||||||
| NC score | -0.003363 (rank : 132) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
ZN167_HUMAN
|
||||||
| NC score | -0.005194 (rank : 133) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P0L1, Q6WL09, Q96FQ2 | Gene names | ZNF167, ZNF448, ZNF64 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 167 (Zinc finger protein 64) (Zinc finger protein 448) (ZFP). | |||||