Please be patient as the page loads
|
KTNA1_HUMAN
|
||||||
| SwissProt Accessions | O75449, Q5TFA8, Q5TFA9, Q86VN2, Q9NU52 | Gene names | KATNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
KTNA1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | O75449, Q5TFA8, Q5TFA9, Q86VN2, Q9NU52 | Gene names | KATNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin). | |||||
|
KTNA1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.997142 (rank : 2) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
|
KATL1_HUMAN
|
||||||
| θ value | 2.40406e-178 (rank : 3) | NC score | 0.995133 (rank : 4) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9BW62 | Gene names | KATNAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
KATL1_MOUSE
|
||||||
| θ value | 2.25013e-176 (rank : 4) | NC score | 0.995159 (rank : 3) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8K0T4 | Gene names | Katnal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
VPS4A_HUMAN
|
||||||
| θ value | 5.17823e-64 (rank : 5) | NC score | 0.957970 (rank : 11) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| θ value | 5.17823e-64 (rank : 6) | NC score | 0.957939 (rank : 12) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
SPAST_HUMAN
|
||||||
| θ value | 6.76295e-64 (rank : 7) | NC score | 0.962764 (rank : 5) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UBP0, Q9UPR9 | Gene names | SPAST, KIAA1083, SPG4 | |||
|
Domain Architecture |

|
|||||
| Description | Spastin. | |||||
|
SPAST_MOUSE
|
||||||
| θ value | 7.47731e-63 (rank : 8) | NC score | 0.961469 (rank : 6) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
VPS4B_MOUSE
|
||||||
| θ value | 7.47731e-63 (rank : 9) | NC score | 0.959559 (rank : 10) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4B_HUMAN
|
||||||
| θ value | 1.07972e-61 (rank : 10) | NC score | 0.959997 (rank : 9) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
ATAD1_HUMAN
|
||||||
| θ value | 5.20228e-48 (rank : 11) | NC score | 0.961443 (rank : 7) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8NBU5, Q6P4B9, Q8N3G1, Q8WYR9, Q969Y3 | Gene names | ATAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
ATAD1_MOUSE
|
||||||
| θ value | 5.20228e-48 (rank : 12) | NC score | 0.961443 (rank : 8) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9D5T0, Q3U8V2, Q9D7A4 | Gene names | Atad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
FIGN_MOUSE
|
||||||
| θ value | 1.15895e-47 (rank : 13) | NC score | 0.928323 (rank : 17) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
FIGN_HUMAN
|
||||||
| θ value | 4.40402e-47 (rank : 14) | NC score | 0.923864 (rank : 22) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
TERA_HUMAN
|
||||||
| θ value | 7.03111e-45 (rank : 15) | NC score | 0.929933 (rank : 13) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P55072, Q969G7 | Gene names | VCP | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
TERA_MOUSE
|
||||||
| θ value | 9.18288e-45 (rank : 16) | NC score | 0.929890 (rank : 14) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q01853 | Gene names | Vcp | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
PRS8_HUMAN
|
||||||
| θ value | 1.2411e-41 (rank : 17) | NC score | 0.927278 (rank : 18) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P62195, O35051, O43208, P47210, P52915, P52916 | Gene names | PSMC5, SUG1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (Thyroid hormone receptor- interacting protein 1) (TRIP1). | |||||
|
PRS8_MOUSE
|
||||||
| θ value | 1.2411e-41 (rank : 18) | NC score | 0.927278 (rank : 19) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P62196, O35051, P47210, P52915, P52916, Q3UL51, Q9CWN5 | Gene names | Psmc5, Sug1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (mSUG1). | |||||
|
PRS10_HUMAN
|
||||||
| θ value | 5.21438e-40 (rank : 19) | NC score | 0.928710 (rank : 15) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P62333, P49719, Q6IBU3, Q92524 | Gene names | PSMC6, SUG2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS10_MOUSE
|
||||||
| θ value | 5.21438e-40 (rank : 20) | NC score | 0.928710 (rank : 16) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P62334, P49719, Q3TKK1, Q810A6, Q92524, Q9CXH9 | Gene names | Psmc6, Sug2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS4_HUMAN
|
||||||
| θ value | 1.51715e-39 (rank : 21) | NC score | 0.923488 (rank : 23) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P62191, P49014, Q03527, Q6IAW0, Q96AZ3 | Gene names | PSMC1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS4_MOUSE
|
||||||
| θ value | 1.51715e-39 (rank : 22) | NC score | 0.923488 (rank : 24) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P62192, P49014, Q03527, Q96AZ3 | Gene names | Psmc1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
NVL_HUMAN
|
||||||
| θ value | 5.76516e-39 (rank : 23) | NC score | 0.919351 (rank : 27) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
NVL_MOUSE
|
||||||
| θ value | 7.52953e-39 (rank : 24) | NC score | 0.919901 (rank : 26) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PRS6A_HUMAN
|
||||||
| θ value | 1.08726e-37 (rank : 25) | NC score | 0.924449 (rank : 21) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P17980, Q96HD3 | Gene names | PSMC3, TBP1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1) (Proteasome subunit P50). | |||||
|
PRS6A_MOUSE
|
||||||
| θ value | 2.42216e-37 (rank : 26) | NC score | 0.924634 (rank : 20) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
|
PEX6_MOUSE
|
||||||
| θ value | 7.04741e-37 (rank : 27) | NC score | 0.914815 (rank : 33) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PEX6_HUMAN
|
||||||
| θ value | 2.67802e-36 (rank : 28) | NC score | 0.915081 (rank : 32) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PEX1_HUMAN
|
||||||
| θ value | 2.96089e-35 (rank : 29) | NC score | 0.922484 (rank : 25) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
YMEL1_MOUSE
|
||||||
| θ value | 1.12514e-34 (rank : 30) | NC score | 0.903856 (rank : 34) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
ATAD2_HUMAN
|
||||||
| θ value | 4.72714e-33 (rank : 31) | NC score | 0.853903 (rank : 43) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
ATAD2_MOUSE
|
||||||
| θ value | 8.06329e-33 (rank : 32) | NC score | 0.871140 (rank : 42) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
AFG31_MOUSE
|
||||||
| θ value | 1.37539e-32 (rank : 33) | NC score | 0.898991 (rank : 36) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q920A7 | Gene names | Afg3l1 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 1 (EC 3.4.24.-). | |||||
|
PRS6B_HUMAN
|
||||||
| θ value | 1.37539e-32 (rank : 34) | NC score | 0.918038 (rank : 30) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P43686, Q96FV5, Q9UBM3, Q9UEX3 | Gene names | PSMC4, TBP7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7). | |||||
|
PRS7_HUMAN
|
||||||
| θ value | 1.79631e-32 (rank : 35) | NC score | 0.918791 (rank : 28) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P35998 | Gene names | PSMC2, MSS1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS7_MOUSE
|
||||||
| θ value | 1.79631e-32 (rank : 36) | NC score | 0.918782 (rank : 29) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P46471, O08531, Q3TGX5, Q9DBA1 | Gene names | Psmc2, Mss1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS6B_MOUSE
|
||||||
| θ value | 6.82597e-32 (rank : 37) | NC score | 0.916862 (rank : 31) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P54775 | Gene names | Psmc4, Tbp7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7) (CIP21). | |||||
|
AFG32_HUMAN
|
||||||
| θ value | 3.3877e-31 (rank : 38) | NC score | 0.893148 (rank : 38) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9Y4W6 | Gene names | AFG3L2 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-) (Paraplegin-like protein). | |||||
|
AFG32_MOUSE
|
||||||
| θ value | 4.42448e-31 (rank : 39) | NC score | 0.892973 (rank : 39) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8JZQ2 | Gene names | Afg3l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-). | |||||
|
YMEL1_HUMAN
|
||||||
| θ value | 7.54701e-31 (rank : 40) | NC score | 0.900325 (rank : 35) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96TA2, Q9H1Q0, Q9UMR9 | Gene names | YME1L1, FTSH1, YME1L | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin- associated metalloprotease) (PAMP). | |||||
|
SPG7_HUMAN
|
||||||
| θ value | 3.74554e-30 (rank : 41) | NC score | 0.898262 (rank : 37) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UQ90, O75756, Q96IB0 | Gene names | SPG7, CAR, CMAR, PGN | |||
|
Domain Architecture |

|
|||||
| Description | Paraplegin (EC 3.4.24.-) (Spastic paraplegia protein 7). | |||||
|
NSF_HUMAN
|
||||||
| θ value | 5.79196e-23 (rank : 42) | NC score | 0.879257 (rank : 40) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P46459, Q9UKZ2 | Gene names | NSF | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein). | |||||
|
NSF_MOUSE
|
||||||
| θ value | 5.79196e-23 (rank : 43) | NC score | 0.878785 (rank : 41) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P46460 | Gene names | Nsf, Skd2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein) (SKD2 protein). | |||||
|
TRP13_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 44) | NC score | 0.717015 (rank : 45) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q15645, O15324 | Gene names | TRIP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13) (Human papillomavirus type 16 E1 protein- binding protein) (HPV16 E1 protein-binding protein) (16E1-BP). | |||||
|
TRP13_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 45) | NC score | 0.718482 (rank : 44) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q3UA06, Q3UQG6, Q9CWW8 | Gene names | Trip13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13). | |||||
|
ATD3B_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 46) | NC score | 0.623496 (rank : 47) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
ATD3A_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 47) | NC score | 0.638767 (rank : 46) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9NVI7, Q8N275, Q96A50 | Gene names | ATAD3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3A. | |||||
|
ATAD3_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 48) | NC score | 0.614536 (rank : 48) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
CLPX_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 49) | NC score | 0.425856 (rank : 52) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JHS4, Q9WVD1 | Gene names | Clpx | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
CLPX_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 50) | NC score | 0.428688 (rank : 51) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
RUVB1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 51) | NC score | 0.324907 (rank : 54) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9Y265, Q9BSX9 | Gene names | RUVBL1, NMP238, TIP49, TIP49A | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (Nuclear matrix protein 238) (NMP 238) (54 kDa erythrocyte cytosolic protein) (ECP-54) (TIP60-associated protein 54-alpha) (TAP54-alpha). | |||||
|
RUVB1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 52) | NC score | 0.324867 (rank : 55) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P60122, O35753 | Gene names | Ruvbl1, Tip49, Tip49a | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (DNA helicase p50). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 0.125558 (rank : 53) | NC score | 0.010860 (rank : 92) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 54) | NC score | 0.010419 (rank : 94) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
FOXM1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 55) | NC score | 0.017424 (rank : 74) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08050, O43258, O43259, O43260, Q4ZGG7, Q9BRL2 | Gene names | FOXM1, FKHL16, HFH11, MPP2, WIN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead-related protein FKHL16) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/fork-head homolog 11) (HFH-11) (Winged helix factor from INS-1 cells) (M-phase phosphoprotein 2) (MPM-2 reactive phosphoprotein 2) (Transcription factor Trident). | |||||
|
IMUP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 56) | NC score | 0.075869 (rank : 68) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9GZP8, Q96H58, Q9HCR4 | Gene names | IMUP, C19orf33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immortalization-up-regulated protein (Hepatocyte growth factor activator inhibitor type 2-related small protein) (HAI-2-related small protein) (H2RSP). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 0.365318 (rank : 57) | NC score | 0.012445 (rank : 86) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
WRIP1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 58) | NC score | 0.230493 (rank : 56) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 59) | NC score | 0.016536 (rank : 75) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
THNSL_MOUSE
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.131636 (rank : 58) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BH55 | Gene names | Thnsl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
THOC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 61) | NC score | 0.025397 (rank : 72) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
WRIP1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 62) | NC score | 0.229009 (rank : 57) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 63) | NC score | 0.016432 (rank : 76) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
BCS1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.565687 (rank : 50) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9CZP5 | Gene names | Bcs1l | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein). | |||||
|
SFRS6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.009501 (rank : 96) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
LONM_HUMAN
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.408500 (rank : 53) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
RFC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.120503 (rank : 61) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
RFC1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.130121 (rank : 59) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.009611 (rank : 95) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
TSH1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.007115 (rank : 104) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.011360 (rank : 91) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BCS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.576281 (rank : 49) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y276, Q7Z2V7 | Gene names | BCS1L, BCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein) (H-BCS1). | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.006672 (rank : 105) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.013903 (rank : 79) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.013575 (rank : 82) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
THNSL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.120059 (rank : 63) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IYQ7, Q5VV21 | Gene names | THNSL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | -0.003015 (rank : 118) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
DEK_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.013156 (rank : 84) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
ENL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.012888 (rank : 85) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
SGOL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.012098 (rank : 87) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5FBB7, Q588H5, Q5FBB4, Q5FBB5, Q5FBB6, Q5FBB8, Q8N579, Q8WVL0, Q9BVA8, Q9H275 | Gene names | SGOL1, SGO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 1 (hSgo1) (Serologically defined breast cancer antigen NY-BR-85). | |||||
|
LYAR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.010858 (rank : 93) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08288, Q9D9X2 | Gene names | Lyar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
M4K4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | -0.005856 (rank : 120) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
MCPH1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.009229 (rank : 100) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TT79, Q8BNI9, Q8C9I7 | Gene names | Mcph1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
RFC4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.104778 (rank : 65) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P35249, Q6FHX7 | Gene names | RFC4 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 4 (Replication factor C 37 kDa subunit) (RF-C 37 kDa subunit) (RFC37) (Activator 1 37 kDa subunit) (A1 37 kDa subunit). | |||||
|
RTKN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.014924 (rank : 78) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C6B2, Q61192, Q8VIG7 | Gene names | Rtkn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rhotekin. | |||||
|
TR150_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.011540 (rank : 90) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y2W1, Q5VTK6 | Gene names | THRAP3, TRAP150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
U2AFM_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.002693 (rank : 112) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
CBLB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.005859 (rank : 108) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3TTA7 | Gene names | Cblb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase CBL-B (EC 6.3.2.-) (Signal transduction protein CBL-B) (SH3-binding protein CBL-B) (Casitas B-lineage lymphoma proto-oncogene b). | |||||
|
CIR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.027201 (rank : 71) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
DYH5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.009284 (rank : 98) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
NAB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.011953 (rank : 88) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13506, O75383, O75384, Q6GTU1, Q9UEV1 | Gene names | NAB1 | |||
|
Domain Architecture |
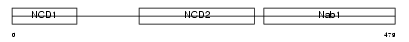
|
|||||
| Description | NGFI-A-binding protein 1 (EGR-1-binding protein 1) (Transcriptional regulatory protein p54). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.011578 (rank : 89) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
RFC4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.093075 (rank : 66) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99J62 | Gene names | Rfc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Replication factor C subunit 4. | |||||
|
SFR12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.013892 (rank : 80) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.007487 (rank : 103) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
AF9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.013287 (rank : 83) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
APC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.007843 (rank : 101) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
BANK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.018804 (rank : 73) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80VH0, Q3U178, Q8BRV6, Q8BRY8 | Gene names | Bank1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell scaffold protein with ankyrin repeats (Protein AVIEF). | |||||
|
BARX2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | -0.000507 (rank : 117) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08686 | Gene names | Barx2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein BarH-like 2. | |||||
|
CTCF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | -0.004878 (rank : 119) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49711 | Gene names | CTCF | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor CTCF (CCCTC-binding factor) (CTCFL paralog) (11-zinc finger protein). | |||||
|
ERCC6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.004944 (rank : 109) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q03468 | Gene names | ERCC6, CSB | |||
|
Domain Architecture |

|
|||||
| Description | DNA excision repair protein ERCC-6 (EC 3.6.1.-) (ATP-dependent helicase ERCC6) (Cockayne syndrome protein CSB). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.007798 (rank : 102) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
PAWR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.006455 (rank : 106) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96IZ0, O75796, Q6FHY9, Q8N700 | Gene names | PAWR, PAR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRKC apoptosis WT1 regulator protein (Prostate apoptosis response 4 protein) (Par-4). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.009435 (rank : 97) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
ACHA6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | -0.000232 (rank : 116) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0W9, Q8K0A7 | Gene names | Chrna6, Nica6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal acetylcholine receptor protein subunit alpha-6 precursor. | |||||
|
DBNL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.001402 (rank : 114) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UJU6, P84070, Q6IAI8, Q96F30, Q96K74, Q9HBN8, Q9NR72 | Gene names | DBNL, CMAP, SH3P7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Drebrin-like protein (SH3 domain-containing protein 7) (Drebrin F) (Cervical SH3P7) (HPK1-interacting protein of 55 kDa) (HIP-55) (Cervical mucin-associated protein). | |||||
|
HELZ_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.003505 (rank : 110) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
KAD6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.107389 (rank : 64) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
LIMA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.013677 (rank : 81) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ERG0, Q9ERG1 | Gene names | Lima1, D15Ertd366e, Eplin | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm) (mEPLIN). | |||||
|
LRRF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.002469 (rank : 113) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.003088 (rank : 111) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
NMDE4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.000628 (rank : 115) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.014966 (rank : 77) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RUVB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.123872 (rank : 60) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y230, Q6PK27, Q9Y361 | Gene names | RUVBL2, TIP48, TIP49B | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (48 kDa TATA box-binding protein-interacting protein) (48 kDa TBP-interacting protein) (TIP49b) (Repressing pontin 52) (Reptin 52) (51 kDa erythrocyte cytosolic protein) (ECP-51) (TIP60-associated protein 54-beta) (TAP54-beta). | |||||
|
RUVB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.120334 (rank : 62) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WTM5 | Gene names | Ruvbl2 | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (p47 protein). | |||||
|
SFRS4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.005995 (rank : 107) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
TR150_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.009258 (rank : 99) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
KAD6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.082095 (rank : 67) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VCP8 | Gene names | Ak6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
RFC5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.057366 (rank : 69) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P40937 | Gene names | RFC5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RFC5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.056612 (rank : 70) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9D0F6 | Gene names | Rfc5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
KTNA1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | O75449, Q5TFA8, Q5TFA9, Q86VN2, Q9NU52 | Gene names | KATNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin). | |||||
|
KTNA1_MOUSE
|
||||||
| NC score | 0.997142 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
|
KATL1_MOUSE
|
||||||
| NC score | 0.995159 (rank : 3) | θ value | 2.25013e-176 (rank : 4) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8K0T4 | Gene names | Katnal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
KATL1_HUMAN
|
||||||
| NC score | 0.995133 (rank : 4) | θ value | 2.40406e-178 (rank : 3) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9BW62 | Gene names | KATNAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
SPAST_HUMAN
|
||||||
| NC score | 0.962764 (rank : 5) | θ value | 6.76295e-64 (rank : 7) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UBP0, Q9UPR9 | Gene names | SPAST, KIAA1083, SPG4 | |||
|
Domain Architecture |

|
|||||
| Description | Spastin. | |||||
|
SPAST_MOUSE
|
||||||
| NC score | 0.961469 (rank : 6) | θ value | 7.47731e-63 (rank : 8) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
ATAD1_HUMAN
|
||||||
| NC score | 0.961443 (rank : 7) | θ value | 5.20228e-48 (rank : 11) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8NBU5, Q6P4B9, Q8N3G1, Q8WYR9, Q969Y3 | Gene names | ATAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
ATAD1_MOUSE
|
||||||
| NC score | 0.961443 (rank : 8) | θ value | 5.20228e-48 (rank : 12) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9D5T0, Q3U8V2, Q9D7A4 | Gene names | Atad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
VPS4B_HUMAN
|
||||||
| NC score | 0.959997 (rank : 9) | θ value | 1.07972e-61 (rank : 10) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4B_MOUSE
|
||||||
| NC score | 0.959559 (rank : 10) | θ value | 7.47731e-63 (rank : 9) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4A_HUMAN
|
||||||
| NC score | 0.957970 (rank : 11) | θ value | 5.17823e-64 (rank : 5) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| NC score | 0.957939 (rank : 12) | θ value | 5.17823e-64 (rank : 6) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
TERA_HUMAN
|
||||||
| NC score | 0.929933 (rank : 13) | θ value | 7.03111e-45 (rank : 15) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P55072, Q969G7 | Gene names | VCP | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
TERA_MOUSE
|
||||||
| NC score | 0.929890 (rank : 14) | θ value | 9.18288e-45 (rank : 16) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q01853 | Gene names | Vcp | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
PRS10_HUMAN
|
||||||
| NC score | 0.928710 (rank : 15) | θ value | 5.21438e-40 (rank : 19) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P62333, P49719, Q6IBU3, Q92524 | Gene names | PSMC6, SUG2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS10_MOUSE
|
||||||
| NC score | 0.928710 (rank : 16) | θ value | 5.21438e-40 (rank : 20) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P62334, P49719, Q3TKK1, Q810A6, Q92524, Q9CXH9 | Gene names | Psmc6, Sug2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
FIGN_MOUSE
|
||||||
| NC score | 0.928323 (rank : 17) | θ value | 1.15895e-47 (rank : 13) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
PRS8_HUMAN
|
||||||
| NC score | 0.927278 (rank : 18) | θ value | 1.2411e-41 (rank : 17) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P62195, O35051, O43208, P47210, P52915, P52916 | Gene names | PSMC5, SUG1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (Thyroid hormone receptor- interacting protein 1) (TRIP1). | |||||
|
PRS8_MOUSE
|
||||||
| NC score | 0.927278 (rank : 19) | θ value | 1.2411e-41 (rank : 18) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P62196, O35051, P47210, P52915, P52916, Q3UL51, Q9CWN5 | Gene names | Psmc5, Sug1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (mSUG1). | |||||
|
PRS6A_MOUSE
|
||||||
| NC score | 0.924634 (rank : 20) | θ value | 2.42216e-37 (rank : 26) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
|
PRS6A_HUMAN
|
||||||
| NC score | 0.924449 (rank : 21) | θ value | 1.08726e-37 (rank : 25) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P17980, Q96HD3 | Gene names | PSMC3, TBP1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1) (Proteasome subunit P50). | |||||
|
FIGN_HUMAN
|
||||||
| NC score | 0.923864 (rank : 22) | θ value | 4.40402e-47 (rank : 14) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
PRS4_HUMAN
|
||||||
| NC score | 0.923488 (rank : 23) | θ value | 1.51715e-39 (rank : 21) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P62191, P49014, Q03527, Q6IAW0, Q96AZ3 | Gene names | PSMC1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS4_MOUSE
|
||||||
| NC score | 0.923488 (rank : 24) | θ value | 1.51715e-39 (rank : 22) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P62192, P49014, Q03527, Q96AZ3 | Gene names | Psmc1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PEX1_HUMAN
|
||||||
| NC score | 0.922484 (rank : 25) | θ value | 2.96089e-35 (rank : 29) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
NVL_MOUSE
|
||||||
| NC score | 0.919901 (rank : 26) | θ value | 7.52953e-39 (rank : 24) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
NVL_HUMAN
|
||||||
| NC score | 0.919351 (rank : 27) | θ value | 5.76516e-39 (rank : 23) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PRS7_HUMAN
|
||||||
| NC score | 0.918791 (rank : 28) | θ value | 1.79631e-32 (rank : 35) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P35998 | Gene names | PSMC2, MSS1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS7_MOUSE
|
||||||
| NC score | 0.918782 (rank : 29) | θ value | 1.79631e-32 (rank : 36) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P46471, O08531, Q3TGX5, Q9DBA1 | Gene names | Psmc2, Mss1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS6B_HUMAN
|
||||||
| NC score | 0.918038 (rank : 30) | θ value | 1.37539e-32 (rank : 34) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P43686, Q96FV5, Q9UBM3, Q9UEX3 | Gene names | PSMC4, TBP7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7). | |||||
|
PRS6B_MOUSE
|
||||||
| NC score | 0.916862 (rank : 31) | θ value | 6.82597e-32 (rank : 37) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P54775 | Gene names | Psmc4, Tbp7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7) (CIP21). | |||||
|
PEX6_HUMAN
|
||||||
| NC score | 0.915081 (rank : 32) | θ value | 2.67802e-36 (rank : 28) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PEX6_MOUSE
|
||||||
| NC score | 0.914815 (rank : 33) | θ value | 7.04741e-37 (rank : 27) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
YMEL1_MOUSE
|
||||||
| NC score | 0.903856 (rank : 34) | θ value | 1.12514e-34 (rank : 30) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
YMEL1_HUMAN
|
||||||
| NC score | 0.900325 (rank : 35) | θ value | 7.54701e-31 (rank : 40) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96TA2, Q9H1Q0, Q9UMR9 | Gene names | YME1L1, FTSH1, YME1L | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin- associated metalloprotease) (PAMP). | |||||
|
AFG31_MOUSE
|
||||||
| NC score | 0.898991 (rank : 36) | θ value | 1.37539e-32 (rank : 33) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q920A7 | Gene names | Afg3l1 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 1 (EC 3.4.24.-). | |||||
|
SPG7_HUMAN
|
||||||
| NC score | 0.898262 (rank : 37) | θ value | 3.74554e-30 (rank : 41) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UQ90, O75756, Q96IB0 | Gene names | SPG7, CAR, CMAR, PGN | |||
|
Domain Architecture |

|
|||||
| Description | Paraplegin (EC 3.4.24.-) (Spastic paraplegia protein 7). | |||||
|
AFG32_HUMAN
|
||||||
| NC score | 0.893148 (rank : 38) | θ value | 3.3877e-31 (rank : 38) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9Y4W6 | Gene names | AFG3L2 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-) (Paraplegin-like protein). | |||||
|
AFG32_MOUSE
|
||||||
| NC score | 0.892973 (rank : 39) | θ value | 4.42448e-31 (rank : 39) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8JZQ2 | Gene names | Afg3l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-). | |||||
|
NSF_HUMAN
|
||||||
| NC score | 0.879257 (rank : 40) | θ value | 5.79196e-23 (rank : 42) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P46459, Q9UKZ2 | Gene names | NSF | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein). | |||||
|
NSF_MOUSE
|
||||||
| NC score | 0.878785 (rank : 41) | θ value | 5.79196e-23 (rank : 43) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P46460 | Gene names | Nsf, Skd2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein) (SKD2 protein). | |||||
|
ATAD2_MOUSE
|
||||||
| NC score | 0.871140 (rank : 42) | θ value | 8.06329e-33 (rank : 32) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
ATAD2_HUMAN
|
||||||
| NC score | 0.853903 (rank : 43) | θ value | 4.72714e-33 (rank : 31) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
TRP13_MOUSE
|
||||||
| NC score | 0.718482 (rank : 44) | θ value | 2.13673e-09 (rank : 45) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q3UA06, Q3UQG6, Q9CWW8 | Gene names | Trip13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13). | |||||
|
TRP13_HUMAN
|
||||||
| NC score | 0.717015 (rank : 45) | θ value | 2.13673e-09 (rank : 44) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q15645, O15324 | Gene names | TRIP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13) (Human papillomavirus type 16 E1 protein- binding protein) (HPV16 E1 protein-binding protein) (16E1-BP). | |||||
|
ATD3A_HUMAN
|
||||||
| NC score | 0.638767 (rank : 46) | θ value | 1.38499e-08 (rank : 47) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9NVI7, Q8N275, Q96A50 | Gene names | ATAD3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3A. | |||||
|
ATD3B_HUMAN
|
||||||
| NC score | 0.623496 (rank : 47) | θ value | 4.76016e-09 (rank : 46) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
ATAD3_MOUSE
|
||||||
| NC score | 0.614536 (rank : 48) | θ value | 2.36244e-08 (rank : 48) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
BCS1_HUMAN
|
||||||
| NC score | 0.576281 (rank : 49) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y276, Q7Z2V7 | Gene names | BCS1L, BCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein) (H-BCS1). | |||||
|
BCS1_MOUSE
|
||||||
| NC score | 0.565687 (rank : 50) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9CZP5 | Gene names | Bcs1l | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein). | |||||
|
CLPX_HUMAN
|
||||||
| NC score | 0.428688 (rank : 51) | θ value | 0.00228821 (rank : 50) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
CLPX_MOUSE
|
||||||
| NC score | 0.425856 (rank : 52) | θ value | 0.00175202 (rank : 49) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JHS4, Q9WVD1 | Gene names | Clpx | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
LONM_HUMAN
|
||||||
| NC score | 0.408500 (rank : 53) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
RUVB1_HUMAN
|
||||||
| NC score | 0.324907 (rank : 54) | θ value | 0.0563607 (rank : 51) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9Y265, Q9BSX9 | Gene names | RUVBL1, NMP238, TIP49, TIP49A | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (Nuclear matrix protein 238) (NMP 238) (54 kDa erythrocyte cytosolic protein) (ECP-54) (TIP60-associated protein 54-alpha) (TAP54-alpha). | |||||
|
RUVB1_MOUSE
|
||||||
| NC score | 0.324867 (rank : 55) | θ value | 0.0563607 (rank : 52) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P60122, O35753 | Gene names | Ruvbl1, Tip49, Tip49a | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (DNA helicase p50). | |||||
|
WRIP1_HUMAN
|
||||||
| NC score | 0.230493 (rank : 56) | θ value | 0.47712 (rank : 58) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
WRIP1_MOUSE
|
||||||
| NC score | 0.229009 (rank : 57) | θ value | 0.62314 (rank : 62) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
THNSL_MOUSE
|
||||||
| NC score | 0.131636 (rank : 58) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BH55 | Gene names | Thnsl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
RFC1_MOUSE
|
||||||
| NC score | 0.130121 (rank : 59) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
RUVB2_HUMAN
|
||||||
| NC score | 0.123872 (rank : 60) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y230, Q6PK27, Q9Y361 | Gene names | RUVBL2, TIP48, TIP49B | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (48 kDa TATA box-binding protein-interacting protein) (48 kDa TBP-interacting protein) (TIP49b) (Repressing pontin 52) (Reptin 52) (51 kDa erythrocyte cytosolic protein) (ECP-51) (TIP60-associated protein 54-beta) (TAP54-beta). | |||||
|
RFC1_HUMAN
|
||||||
| NC score | 0.120503 (rank : 61) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
RUVB2_MOUSE
|
||||||
| NC score | 0.120334 (rank : 62) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WTM5 | Gene names | Ruvbl2 | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (p47 protein). | |||||
|
THNSL_HUMAN
|
||||||
| NC score | 0.120059 (rank : 63) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IYQ7, Q5VV21 | Gene names | THNSL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
KAD6_HUMAN
|
||||||
| NC score | 0.107389 (rank : 64) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
RFC4_HUMAN
|
||||||
| NC score | 0.104778 (rank : 65) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P35249, Q6FHX7 | Gene names | RFC4 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 4 (Replication factor C 37 kDa subunit) (RF-C 37 kDa subunit) (RFC37) (Activator 1 37 kDa subunit) (A1 37 kDa subunit). | |||||
|
RFC4_MOUSE
|
||||||
| NC score | 0.093075 (rank : 66) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99J62 | Gene names | Rfc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Replication factor C subunit 4. | |||||
|
KAD6_MOUSE
|
||||||
| NC score | 0.082095 (rank : 67) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VCP8 | Gene names | Ak6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
IMUP_HUMAN
|
||||||
| NC score | 0.075869 (rank : 68) | θ value | 0.279714 (rank : 56) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9GZP8, Q96H58, Q9HCR4 | Gene names | IMUP, C19orf33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immortalization-up-regulated protein (Hepatocyte growth factor activator inhibitor type 2-related small protein) (HAI-2-related small protein) (H2RSP). | |||||
|
RFC5_HUMAN
|
||||||
| NC score | 0.057366 (rank : 69) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P40937 | Gene names | RFC5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RFC5_MOUSE
|
||||||
| NC score | 0.056612 (rank : 70) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9D0F6 | Gene names | Rfc5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.027201 (rank : 71) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
THOC2_HUMAN
|
||||||
| NC score | 0.025397 (rank : 72) | θ value | 0.62314 (rank : 61) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
BANK1_MOUSE
|
||||||
| NC score | 0.018804 (rank : 73) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80VH0, Q3U178, Q8BRV6, Q8BRY8 | Gene names | Bank1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell scaffold protein with ankyrin repeats (Protein AVIEF). | |||||
|
FOXM1_HUMAN
|
||||||
| NC score | 0.017424 (rank : 74) | θ value | 0.21417 (rank : 55) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08050, O43258, O43259, O43260, Q4ZGG7, Q9BRL2 | Gene names | FOXM1, FKHL16, HFH11, MPP2, WIN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead-related protein FKHL16) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/fork-head homolog 11) (HFH-11) (Winged helix factor from INS-1 cells) (M-phase phosphoprotein 2) (MPM-2 reactive phosphoprotein 2) (Transcription factor Trident). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.016536 (rank : 75) | θ value | 0.62314 (rank : 59) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.016432 (rank : 76) | θ value | 0.813845 (rank : 63) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.014966 (rank : 77) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RTKN_MOUSE
|
||||||
| NC score | 0.014924 (rank : 78) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C6B2, Q61192, Q8VIG7 | Gene names | Rtkn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rhotekin. | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.013903 (rank : 79) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.013892 (rank : 80) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
LIMA1_MOUSE
|
||||||
| NC score | 0.013677 (rank : 81) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ERG0, Q9ERG1 | Gene names | Lima1, D15Ertd366e, Eplin | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm) (mEPLIN). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.013575 (rank : 82) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
AF9_HUMAN
|
||||||
| NC score | 0.013287 (rank : 83) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
DEK_HUMAN
|
||||||
| NC score | 0.013156 (rank : 84) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.012888 (rank : 85) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.012445 (rank : 86) | θ value | 0.365318 (rank : 57) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
SGOL1_HUMAN
|
||||||
| NC score | 0.012098 (rank : 87) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5FBB7, Q588H5, Q5FBB4, Q5FBB5, Q5FBB6, Q5FBB8, Q8N579, Q8WVL0, Q9BVA8, Q9H275 | Gene names | SGOL1, SGO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 1 (hSgo1) (Serologically defined breast cancer antigen NY-BR-85). | |||||
|
NAB1_HUMAN
|
||||||
| NC score | 0.011953 (rank : 88) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13506, O75383, O75384, Q6GTU1, Q9UEV1 | Gene names | NAB1 | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 1 (EGR-1-binding protein 1) (Transcriptional regulatory protein p54). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.011578 (rank : 89) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
TR150_HUMAN
|
||||||
| NC score | 0.011540 (rank : 90) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y2W1, Q5VTK6 | Gene names | THRAP3, TRAP150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.011360 (rank : 91) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.010860 (rank : 92) | θ value | 0.125558 (rank : 53) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
LYAR_MOUSE
|
||||||
| NC score | 0.010858 (rank : 93) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08288, Q9D9X2 | Gene names | Lyar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.010419 (rank : 94) | θ value | 0.163984 (rank : 54) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.009611 (rank : 95) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
SFRS6_HUMAN
|
||||||
| NC score | 0.009501 (rank : 96) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.009435 (rank : 97) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
DYH5_HUMAN
|
||||||
| NC score | 0.009284 (rank : 98) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
TR150_MOUSE
|
||||||
| NC score | 0.009258 (rank : 99) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
MCPH1_MOUSE
|
||||||
| NC score | 0.009229 (rank : 100) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TT79, Q8BNI9, Q8C9I7 | Gene names | Mcph1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.007843 (rank : 101) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.007798 (rank : 102) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.007487 (rank : 103) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
TSH1_MOUSE
|
||||||
| NC score | 0.007115 (rank : 104) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.006672 (rank : 105) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
PAWR_HUMAN
|
||||||
| NC score | 0.006455 (rank : 106) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96IZ0, O75796, Q6FHY9, Q8N700 | Gene names | PAWR, PAR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRKC apoptosis WT1 regulator protein (Prostate apoptosis response 4 protein) (Par-4). | |||||
|
SFRS4_HUMAN
|
||||||
| NC score | 0.005995 (rank : 107) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
CBLB_MOUSE
|
||||||
| NC score | 0.005859 (rank : 108) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3TTA7 | Gene names | Cblb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase CBL-B (EC 6.3.2.-) (Signal transduction protein CBL-B) (SH3-binding protein CBL-B) (Casitas B-lineage lymphoma proto-oncogene b). | |||||
|
ERCC6_HUMAN
|
||||||
| NC score | 0.004944 (rank : 109) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q03468 | Gene names | ERCC6, CSB | |||
|
Domain Architecture |

|
|||||
| Description | DNA excision repair protein ERCC-6 (EC 3.6.1.-) (ATP-dependent helicase ERCC6) (Cockayne syndrome protein CSB). | |||||
|
HELZ_HUMAN
|
||||||
| NC score | 0.003505 (rank : 110) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.003088 (rank : 111) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
U2AFM_HUMAN
|
||||||
| NC score | 0.002693 (rank : 112) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
LRRF1_MOUSE
|
||||||
| NC score | 0.002469 (rank : 113) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
DBNL_HUMAN
|
||||||
| NC score | 0.001402 (rank : 114) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UJU6, P84070, Q6IAI8, Q96F30, Q96K74, Q9HBN8, Q9NR72 | Gene names | DBNL, CMAP, SH3P7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Drebrin-like protein (SH3 domain-containing protein 7) (Drebrin F) (Cervical SH3P7) (HPK1-interacting protein of 55 kDa) (HIP-55) (Cervical mucin-associated protein). | |||||
|
NMDE4_HUMAN
|
||||||
| NC score | 0.000628 (rank : 115) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
|
ACHA6_MOUSE
|
||||||
| NC score | -0.000232 (rank : 116) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0W9, Q8K0A7 | Gene names | Chrna6, Nica6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal acetylcholine receptor protein subunit alpha-6 precursor. | |||||
|
BARX2_MOUSE
|
||||||
| NC score | -0.000507 (rank : 117) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08686 | Gene names | Barx2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein BarH-like 2. | |||||
|
ANR26_HUMAN
|
||||||
| NC score | -0.003015 (rank : 118) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CTCF_HUMAN
|
||||||
| NC score | -0.004878 (rank : 119) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49711 | Gene names | CTCF | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor CTCF (CCCTC-binding factor) (CTCFL paralog) (11-zinc finger protein). | |||||
|
M4K4_HUMAN
|
||||||
| NC score | -0.005856 (rank : 120) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||