Please be patient as the page loads
|
PRS8_MOUSE
|
||||||
| SwissProt Accessions | P62196, O35051, P47210, P52915, P52916, Q3UL51, Q9CWN5 | Gene names | Psmc5, Sug1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (mSUG1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PRS8_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | P62195, O35051, O43208, P47210, P52915, P52916 | Gene names | PSMC5, SUG1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (Thyroid hormone receptor- interacting protein 1) (TRIP1). | |||||
|
PRS8_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 2) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | P62196, O35051, P47210, P52915, P52916, Q3UL51, Q9CWN5 | Gene names | Psmc5, Sug1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (mSUG1). | |||||
|
PRS4_HUMAN
|
||||||
| θ value | 3.32542e-95 (rank : 3) | NC score | 0.989284 (rank : 3) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P62191, P49014, Q03527, Q6IAW0, Q96AZ3 | Gene names | PSMC1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS4_MOUSE
|
||||||
| θ value | 3.32542e-95 (rank : 4) | NC score | 0.989284 (rank : 4) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P62192, P49014, Q03527, Q96AZ3 | Gene names | Psmc1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS10_HUMAN
|
||||||
| θ value | 1.30768e-91 (rank : 5) | NC score | 0.988802 (rank : 5) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P62333, P49719, Q6IBU3, Q92524 | Gene names | PSMC6, SUG2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS10_MOUSE
|
||||||
| θ value | 1.30768e-91 (rank : 6) | NC score | 0.988802 (rank : 6) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P62334, P49719, Q3TKK1, Q810A6, Q92524, Q9CXH9 | Gene names | Psmc6, Sug2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS7_HUMAN
|
||||||
| θ value | 1.70789e-91 (rank : 7) | NC score | 0.988194 (rank : 7) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P35998 | Gene names | PSMC2, MSS1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS7_MOUSE
|
||||||
| θ value | 1.70789e-91 (rank : 8) | NC score | 0.988182 (rank : 8) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P46471, O08531, Q3TGX5, Q9DBA1 | Gene names | Psmc2, Mss1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS6A_HUMAN
|
||||||
| θ value | 2.16048e-86 (rank : 9) | NC score | 0.985212 (rank : 10) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P17980, Q96HD3 | Gene names | PSMC3, TBP1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1) (Proteasome subunit P50). | |||||
|
PRS6A_MOUSE
|
||||||
| θ value | 1.40038e-85 (rank : 10) | NC score | 0.985299 (rank : 9) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
|
PRS6B_HUMAN
|
||||||
| θ value | 5.50685e-82 (rank : 11) | NC score | 0.983677 (rank : 11) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P43686, Q96FV5, Q9UBM3, Q9UEX3 | Gene names | PSMC4, TBP7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7). | |||||
|
PRS6B_MOUSE
|
||||||
| θ value | 3.94648e-80 (rank : 12) | NC score | 0.983187 (rank : 12) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P54775 | Gene names | Psmc4, Tbp7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7) (CIP21). | |||||
|
AFG32_HUMAN
|
||||||
| θ value | 4.39379e-55 (rank : 13) | NC score | 0.948406 (rank : 18) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9Y4W6 | Gene names | AFG3L2 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-) (Paraplegin-like protein). | |||||
|
AFG32_MOUSE
|
||||||
| θ value | 7.49467e-55 (rank : 14) | NC score | 0.948189 (rank : 19) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8JZQ2 | Gene names | Afg3l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-). | |||||
|
AFG31_MOUSE
|
||||||
| θ value | 1.41344e-53 (rank : 15) | NC score | 0.951936 (rank : 15) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q920A7 | Gene names | Afg3l1 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 1 (EC 3.4.24.-). | |||||
|
TERA_HUMAN
|
||||||
| θ value | 6.56568e-51 (rank : 16) | NC score | 0.952650 (rank : 13) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P55072, Q969G7 | Gene names | VCP | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
TERA_MOUSE
|
||||||
| θ value | 6.56568e-51 (rank : 17) | NC score | 0.952621 (rank : 14) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q01853 | Gene names | Vcp | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
SPG7_HUMAN
|
||||||
| θ value | 3.37202e-47 (rank : 18) | NC score | 0.950531 (rank : 16) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UQ90, O75756, Q96IB0 | Gene names | SPG7, CAR, CMAR, PGN | |||
|
Domain Architecture |

|
|||||
| Description | Paraplegin (EC 3.4.24.-) (Spastic paraplegia protein 7). | |||||
|
PEX6_HUMAN
|
||||||
| θ value | 2.18568e-46 (rank : 19) | NC score | 0.938358 (rank : 26) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
YMEL1_MOUSE
|
||||||
| θ value | 3.72821e-46 (rank : 20) | NC score | 0.943618 (rank : 20) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
YMEL1_HUMAN
|
||||||
| θ value | 6.35935e-46 (rank : 21) | NC score | 0.940047 (rank : 24) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96TA2, Q9H1Q0, Q9UMR9 | Gene names | YME1L1, FTSH1, YME1L | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin- associated metalloprotease) (PAMP). | |||||
|
PEX6_MOUSE
|
||||||
| θ value | 3.15612e-45 (rank : 22) | NC score | 0.938122 (rank : 27) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PEX1_HUMAN
|
||||||
| θ value | 2.04573e-44 (rank : 23) | NC score | 0.949876 (rank : 17) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
ATAD2_HUMAN
|
||||||
| θ value | 2.67181e-44 (rank : 24) | NC score | 0.879506 (rank : 41) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
ATAD2_MOUSE
|
||||||
| θ value | 1.73182e-43 (rank : 25) | NC score | 0.896276 (rank : 40) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
KTNA1_HUMAN
|
||||||
| θ value | 1.2411e-41 (rank : 26) | NC score | 0.927278 (rank : 30) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O75449, Q5TFA8, Q5TFA9, Q86VN2, Q9NU52 | Gene names | KATNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin). | |||||
|
KTNA1_MOUSE
|
||||||
| θ value | 2.76489e-41 (rank : 27) | NC score | 0.925077 (rank : 33) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
|
KATL1_HUMAN
|
||||||
| θ value | 3.61106e-41 (rank : 28) | NC score | 0.930187 (rank : 28) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9BW62 | Gene names | KATNAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
NVL_HUMAN
|
||||||
| θ value | 3.61106e-41 (rank : 29) | NC score | 0.939766 (rank : 25) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
NVL_MOUSE
|
||||||
| θ value | 1.05066e-40 (rank : 30) | NC score | 0.940233 (rank : 23) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
KATL1_MOUSE
|
||||||
| θ value | 1.51715e-39 (rank : 31) | NC score | 0.929876 (rank : 29) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8K0T4 | Gene names | Katnal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
NSF_HUMAN
|
||||||
| θ value | 2.05049e-36 (rank : 32) | NC score | 0.926538 (rank : 32) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P46459, Q9UKZ2 | Gene names | NSF | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein). | |||||
|
NSF_MOUSE
|
||||||
| θ value | 3.4976e-36 (rank : 33) | NC score | 0.926847 (rank : 31) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P46460 | Gene names | Nsf, Skd2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein) (SKD2 protein). | |||||
|
SPAST_HUMAN
|
||||||
| θ value | 1.12514e-34 (rank : 34) | NC score | 0.920727 (rank : 34) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UBP0, Q9UPR9 | Gene names | SPAST, KIAA1083, SPG4 | |||
|
Domain Architecture |

|
|||||
| Description | Spastin. | |||||
|
SPAST_MOUSE
|
||||||
| θ value | 1.12514e-34 (rank : 35) | NC score | 0.919217 (rank : 35) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
ATAD1_HUMAN
|
||||||
| θ value | 1.46948e-34 (rank : 36) | NC score | 0.940978 (rank : 21) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8NBU5, Q6P4B9, Q8N3G1, Q8WYR9, Q969Y3 | Gene names | ATAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
ATAD1_MOUSE
|
||||||
| θ value | 1.46948e-34 (rank : 37) | NC score | 0.940978 (rank : 22) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9D5T0, Q3U8V2, Q9D7A4 | Gene names | Atad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
VPS4A_HUMAN
|
||||||
| θ value | 2.77131e-33 (rank : 38) | NC score | 0.909272 (rank : 38) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| θ value | 2.77131e-33 (rank : 39) | NC score | 0.909253 (rank : 39) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
VPS4B_HUMAN
|
||||||
| θ value | 4.00176e-32 (rank : 40) | NC score | 0.911554 (rank : 36) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4B_MOUSE
|
||||||
| θ value | 1.52067e-31 (rank : 41) | NC score | 0.911029 (rank : 37) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
FIGN_MOUSE
|
||||||
| θ value | 3.87602e-27 (rank : 42) | NC score | 0.874216 (rank : 42) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
FIGN_HUMAN
|
||||||
| θ value | 1.47289e-26 (rank : 43) | NC score | 0.868451 (rank : 43) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
BCS1_MOUSE
|
||||||
| θ value | 1.2105e-12 (rank : 44) | NC score | 0.710968 (rank : 45) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9CZP5 | Gene names | Bcs1l | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein). | |||||
|
BCS1_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 45) | NC score | 0.718335 (rank : 44) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y276, Q7Z2V7 | Gene names | BCS1L, BCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein) (H-BCS1). | |||||
|
ATD3B_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 46) | NC score | 0.629192 (rank : 49) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
ATAD3_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 47) | NC score | 0.619934 (rank : 50) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
ATD3A_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 48) | NC score | 0.637878 (rank : 48) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9NVI7, Q8N275, Q96A50 | Gene names | ATAD3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3A. | |||||
|
TRP13_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 49) | NC score | 0.710382 (rank : 46) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q3UA06, Q3UQG6, Q9CWW8 | Gene names | Trip13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13). | |||||
|
TRP13_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 50) | NC score | 0.706895 (rank : 47) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q15645, O15324 | Gene names | TRIP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13) (Human papillomavirus type 16 E1 protein- binding protein) (HPV16 E1 protein-binding protein) (16E1-BP). | |||||
|
CLPX_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 51) | NC score | 0.459857 (rank : 51) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
CLPX_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 52) | NC score | 0.457928 (rank : 52) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JHS4, Q9WVD1 | Gene names | Clpx | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
LONM_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 53) | NC score | 0.440437 (rank : 53) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
RENT1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 54) | NC score | 0.045190 (rank : 70) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9EPU0, Q99PR4 | Gene names | Upf1, Rent1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (mUpf1). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 55) | NC score | 0.011075 (rank : 107) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
RENT1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 56) | NC score | 0.043742 (rank : 71) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92900, O00239, O43343, Q86Z25, Q92842 | Gene names | UPF1, KIAA0221, RENT1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (hUpf1). | |||||
|
RUVB1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 57) | NC score | 0.318985 (rank : 54) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y265, Q9BSX9 | Gene names | RUVBL1, NMP238, TIP49, TIP49A | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (Nuclear matrix protein 238) (NMP 238) (54 kDa erythrocyte cytosolic protein) (ECP-54) (TIP60-associated protein 54-alpha) (TAP54-alpha). | |||||
|
RUVB1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 58) | NC score | 0.318947 (rank : 55) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P60122, O35753 | Gene names | Ruvbl1, Tip49, Tip49a | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (DNA helicase p50). | |||||
|
RFC1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 59) | NC score | 0.130389 (rank : 58) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
K2C4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 60) | NC score | 0.031551 (rank : 73) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P19013, Q6GTR8, Q96LA7, Q9BTL1 | Gene names | KRT4, CYK4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 4 (Cytokeratin-4) (CK-4) (Keratin-4) (K4). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 0.365318 (rank : 61) | NC score | 0.013726 (rank : 103) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
RFC1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 62) | NC score | 0.115106 (rank : 63) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
TBCD4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 63) | NC score | 0.013662 (rank : 104) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60343, Q5W0B9 | Gene names | TBC1D4, KIAA0603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 4 (Akt substrate of 160 kDa) (AS160). | |||||
|
CING_HUMAN
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.009311 (rank : 109) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
K2C8_MOUSE
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.027670 (rank : 97) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 576 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P11679, Q3KQK5, Q3TGI1, Q3TJE1, Q3TKY7, Q61463, Q61518, Q61519 | Gene names | Krt8, Krt2-8 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8) (Cytokeratin endo A). | |||||
|
WRIP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.235981 (rank : 56) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
K22E_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.028855 (rank : 83) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 515 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35908 | Gene names | KRT2A, KRT2E | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 2 epidermal (Cytokeratin-2e) (K2e) (CK 2e). | |||||
|
K2C1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.027957 (rank : 94) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P04264, Q14720, Q6GSJ0, Q9H298 | Gene names | KRT1, KRTA | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin) (Hair alpha protein). | |||||
|
K2C4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.028371 (rank : 91) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P07744, Q6P3F5 | Gene names | Krt4, Krt2-4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 4 (Cytokeratin-4) (CK-4) (Keratin-4) (K4) (Cytoskeletal 57 kDa keratin). | |||||
|
KRHB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.029038 (rank : 80) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14533, Q14846, Q16274, Q8WU52, Q9BR74 | Gene names | KRTHB1, MLN137 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cuticular Hb1 (Hair keratin, type II Hb1) (ghHKb1) (ghHb1) (MLN 137). | |||||
|
KRHB3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.029173 (rank : 79) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P78385, Q9NSB3 | Gene names | KRTHB3 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cuticular Hb3 (Hair keratin, type II Hb3). | |||||
|
KRHB6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.029026 (rank : 81) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43790, P78387 | Gene names | KRTHB6 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cuticular Hb6 (Hair keratin, type II Hb6) (ghHb6). | |||||
|
M10L1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.024025 (rank : 100) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99MV5, Q7TPA9, Q8C3W0, Q924C2 | Gene names | Mov10l1, Champ | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase Mov10l1 (EC 3.6.1.-) (Moloney leukemia virus 10-like protein 1) (MOV10-like 1) (Cardiac helicase activated by MEF2 protein) (Cardiac-specific RNA helicase). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.043216 (rank : 72) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
RUVB2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.126406 (rank : 59) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y230, Q6PK27, Q9Y361 | Gene names | RUVBL2, TIP48, TIP49B | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (48 kDa TATA box-binding protein-interacting protein) (48 kDa TBP-interacting protein) (TIP49b) (Repressing pontin 52) (Reptin 52) (51 kDa erythrocyte cytosolic protein) (ECP-51) (TIP60-associated protein 54-beta) (TAP54-beta). | |||||
|
RUVB2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.123826 (rank : 60) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9WTM5 | Gene names | Ruvbl2 | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (p47 protein). | |||||
|
THNSL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.115815 (rank : 62) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IYQ7, Q5VV21 | Gene names | THNSL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
THNSL_MOUSE
|
||||||
| θ value | 1.38821 (rank : 78) | NC score | 0.118146 (rank : 61) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BH55 | Gene names | Thnsl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
K2C1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.029889 (rank : 76) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P04104 | Gene names | Krt1, Krt2-1 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin). | |||||
|
TXLNG_MOUSE
|
||||||
| θ value | 1.81305 (rank : 80) | NC score | 0.018432 (rank : 101) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BHN1, Q8BP11 | Gene names | Txlng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin. | |||||
|
WRIP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.232252 (rank : 57) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
IMA7_MOUSE
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.008086 (rank : 111) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35345, Q9CVP9 | Gene names | Kpna6, Kpna5 | |||
|
Domain Architecture |
|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6 subunit) (Importin alpha S2). | |||||
|
K2C1B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.028934 (rank : 82) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6IFZ6 | Gene names | Krt1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 1b (Type II keratin Kb39) (Embryonic type II keratin-1). | |||||
|
KRHB4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.029890 (rank : 75) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NSB2 | Gene names | KRTHB4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb4 (Hair keratin, type II Hb4). | |||||
|
KRHB5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.029305 (rank : 77) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P78386, Q9NSB1 | Gene names | KRTHB5 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cuticular Hb5 (Hair keratin, type II Hb5). | |||||
|
K2C6A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.028724 (rank : 84) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P50446, Q9Z332 | Gene names | Krt6a, Ker2, Krt2-6, Krt2-6a, Krt6 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6A (Cytokeratin-6A) (CK 6A) (K6a keratin) (Keratin-6 alpha) (mK6-alpha). | |||||
|
K2C6B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.028627 (rank : 86) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Z331 | Gene names | Krt6b, K6-beta, Krt2-6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 6B (Cytokeratin-6B) (CK 6B) (K6b keratin) (Keratin-6 beta) (mK6-beta). | |||||
|
PIBF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.008013 (rank : 112) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
TPR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.011068 (rank : 108) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
CEP63_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.006705 (rank : 118) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
CR034_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.011148 (rank : 106) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5BJE1, Q6ZP67, Q6ZU20 | Gene names | C18orf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34. | |||||
|
K2C6A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.028598 (rank : 87) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P02538, Q6NT67, Q96CL4 | Gene names | KRT6A, K6A | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6A (Cytokeratin-6A) (CK 6A) (K6a keratin). | |||||
|
K2C6B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.028558 (rank : 88) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P04259, P48669 | Gene names | KRT6B, K6B | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6B (Cytokeratin-6B) (CK 6B) (K6b keratin). | |||||
|
K2C6C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.028538 (rank : 89) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P48666 | Gene names | KRT6C | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6C (Cytokeratin-6C) (CK 6C) (K6c keratin). | |||||
|
K2C6E_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.028509 (rank : 90) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P48668, Q7RTN9 | Gene names | KRT6E | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6E (Cytokeratin-6E) (CK 6E) (K6e keratin) (Keratin K6h). | |||||
|
K2C8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.026566 (rank : 98) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P05787, Q14099, Q14716, Q14717, Q53GJ0, Q6DHW5, Q6GMY0, Q96J60 | Gene names | KRT8, CYK8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8). | |||||
|
TXLNG_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.016718 (rank : 102) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NUQ3, Q9P0X1 | Gene names | TXLNG, CXorf15, LSR5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin (Lipopolysaccharide-specific response protein 5). | |||||
|
HOOK3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.006919 (rank : 115) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
K22O_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.028121 (rank : 93) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q01546, Q7Z795 | Gene names | KRT76, KRT2B, KRT2P | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 2 oral (Cytokeratin-2P) (K2P) (CK 2P) (Keratin 76). | |||||
|
K2C5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.027853 (rank : 96) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P13647, Q6PI71, Q6UBJ0, Q8TA91 | Gene names | KRT5 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 5 (Cytokeratin-5) (CK-5) (Keratin-5) (K5) (58 kDa cytokeratin). | |||||
|
K2C5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.027945 (rank : 95) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q922U2, Q920F2 | Gene names | Krt5, Krt2-5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 5 (Cytokeratin-5) (CK-5) (Keratin-5) (K5). | |||||
|
KRHB5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.029287 (rank : 78) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Z2T6, Q8CE83, Q9D7M4 | Gene names | Krthb5, Krt2-18 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cuticular Hb5 (Hair keratin, type II Hb5). | |||||
|
KRHB6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.028652 (rank : 85) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97861, Q3TTV9 | Gene names | Krthb6, Krt2-10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cuticular Hb6 (Hair keratin, type II Hb6). | |||||
|
MOV10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.030479 (rank : 74) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HCE1, Q8TEF0, Q9BSY3, Q9BUJ9 | Gene names | MOV10, KIAA1631 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase MOV-10 (EC 3.6.1.-) (Moloney leukemia virus 10 protein). | |||||
|
MOV10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.025986 (rank : 99) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P23249, Q9DC64 | Gene names | Mov10, Gb110 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase MOV-10 (EC 3.6.1.-) (Moloney leukemia virus 10 protein). | |||||
|
BICD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.006903 (rank : 116) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.005187 (rank : 121) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
RGPD8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.005719 (rank : 119) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99666 | Gene names | RGPD8, RANBP2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RANBP2-like and GRIP domain-containing protein 8 (Ran-binding protein 2-like 1) (RanBP2L1) (Sperm membrane protein BS-63). | |||||
|
RT15_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.013125 (rank : 105) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DC71, Q7TMG9, Q8C7F1, Q9CVE0, Q9CWX1 | Gene names | Mrps15, Rpms15 | |||
|
Domain Architecture |
|
|||||
| Description | 28S ribosomal protein S15, mitochondrial precursor (S15mt) (MRP-S15). | |||||
|
COG3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.007143 (rank : 114) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
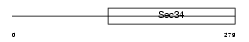
| SwissProt Accessions | Q96JB2, Q5VT70, Q8IXX4, Q9BZ92 | Gene names | COG3, SEC34 | |||
|
Domain Architecture |
|
|||||
| Description | Conserved oligomeric Golgi complex component 3 (Vesicle docking protein SEC34 homolog) (p94). | |||||
|
GCP6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.006850 (rank : 117) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96RT7, Q5JZ80, Q6PJ40, Q86YE9, Q9BY91, Q9UGX3, Q9UGX4 | Gene names | TUBGCP6, GCP6, KIAA1669 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-tubulin complex component 6 (GCP-6). | |||||
|
IQGA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.004419 (rank : 123) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86VI3 | Gene names | IQGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP3. | |||||
|
KRHB4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.028131 (rank : 92) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99M73 | Gene names | Krthb4, Krt2-16 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb4 (Hair keratin, type II Hb4) (65 kDa type II keratin). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.005124 (rank : 122) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.005543 (rank : 120) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
NUP88_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.007442 (rank : 113) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CEC0, Q80Z13, Q8K090 | Gene names | Nup88 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore complex protein Nup88 (Nucleoporin Nup88) (88 kDa nuclear pore complex protein). | |||||
|
RBP22_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.008428 (rank : 110) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q53T03, Q53QN2, Q59GM7 | Gene names | RANBP2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 2 (RanBP2L2). | |||||
|
KAD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.090955 (rank : 65) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
KAD6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.069932 (rank : 67) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VCP8 | Gene names | Ak6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
RFC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.095602 (rank : 64) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P35249, Q6FHX7 | Gene names | RFC4 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 4 (Replication factor C 37 kDa subunit) (RF-C 37 kDa subunit) (RFC37) (Activator 1 37 kDa subunit) (A1 37 kDa subunit). | |||||
|
RFC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.084139 (rank : 66) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99J62 | Gene names | Rfc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Replication factor C subunit 4. | |||||
|
RFC5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.064481 (rank : 69) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P40937 | Gene names | RFC5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RFC5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.065920 (rank : 68) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9D0F6 | Gene names | Rfc5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
PRS8_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | P62195, O35051, O43208, P47210, P52915, P52916 | Gene names | PSMC5, SUG1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (Thyroid hormone receptor- interacting protein 1) (TRIP1). | |||||
|
PRS8_MOUSE
|
||||||
| NC score | 0.999962 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | P62196, O35051, P47210, P52915, P52916, Q3UL51, Q9CWN5 | Gene names | Psmc5, Sug1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (mSUG1). | |||||
|
PRS4_HUMAN
|
||||||
| NC score | 0.989284 (rank : 3) | θ value | 3.32542e-95 (rank : 3) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P62191, P49014, Q03527, Q6IAW0, Q96AZ3 | Gene names | PSMC1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS4_MOUSE
|
||||||
| NC score | 0.989284 (rank : 4) | θ value | 3.32542e-95 (rank : 4) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P62192, P49014, Q03527, Q96AZ3 | Gene names | Psmc1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS10_HUMAN
|
||||||
| NC score | 0.988802 (rank : 5) | θ value | 1.30768e-91 (rank : 5) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P62333, P49719, Q6IBU3, Q92524 | Gene names | PSMC6, SUG2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS10_MOUSE
|
||||||
| NC score | 0.988802 (rank : 6) | θ value | 1.30768e-91 (rank : 6) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P62334, P49719, Q3TKK1, Q810A6, Q92524, Q9CXH9 | Gene names | Psmc6, Sug2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS7_HUMAN
|
||||||
| NC score | 0.988194 (rank : 7) | θ value | 1.70789e-91 (rank : 7) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P35998 | Gene names | PSMC2, MSS1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS7_MOUSE
|
||||||
| NC score | 0.988182 (rank : 8) | θ value | 1.70789e-91 (rank : 8) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P46471, O08531, Q3TGX5, Q9DBA1 | Gene names | Psmc2, Mss1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS6A_MOUSE
|
||||||
| NC score | 0.985299 (rank : 9) | θ value | 1.40038e-85 (rank : 10) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
|
PRS6A_HUMAN
|
||||||
| NC score | 0.985212 (rank : 10) | θ value | 2.16048e-86 (rank : 9) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P17980, Q96HD3 | Gene names | PSMC3, TBP1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1) (Proteasome subunit P50). | |||||
|
PRS6B_HUMAN
|
||||||
| NC score | 0.983677 (rank : 11) | θ value | 5.50685e-82 (rank : 11) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P43686, Q96FV5, Q9UBM3, Q9UEX3 | Gene names | PSMC4, TBP7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7). | |||||
|
PRS6B_MOUSE
|
||||||
| NC score | 0.983187 (rank : 12) | θ value | 3.94648e-80 (rank : 12) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P54775 | Gene names | Psmc4, Tbp7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7) (CIP21). | |||||
|
TERA_HUMAN
|
||||||
| NC score | 0.952650 (rank : 13) | θ value | 6.56568e-51 (rank : 16) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P55072, Q969G7 | Gene names | VCP | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
TERA_MOUSE
|
||||||
| NC score | 0.952621 (rank : 14) | θ value | 6.56568e-51 (rank : 17) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q01853 | Gene names | Vcp | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
AFG31_MOUSE
|
||||||
| NC score | 0.951936 (rank : 15) | θ value | 1.41344e-53 (rank : 15) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q920A7 | Gene names | Afg3l1 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 1 (EC 3.4.24.-). | |||||
|
SPG7_HUMAN
|
||||||
| NC score | 0.950531 (rank : 16) | θ value | 3.37202e-47 (rank : 18) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UQ90, O75756, Q96IB0 | Gene names | SPG7, CAR, CMAR, PGN | |||
|
Domain Architecture |

|
|||||
| Description | Paraplegin (EC 3.4.24.-) (Spastic paraplegia protein 7). | |||||
|
PEX1_HUMAN
|
||||||
| NC score | 0.949876 (rank : 17) | θ value | 2.04573e-44 (rank : 23) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
AFG32_HUMAN
|
||||||
| NC score | 0.948406 (rank : 18) | θ value | 4.39379e-55 (rank : 13) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9Y4W6 | Gene names | AFG3L2 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-) (Paraplegin-like protein). | |||||
|
AFG32_MOUSE
|
||||||
| NC score | 0.948189 (rank : 19) | θ value | 7.49467e-55 (rank : 14) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8JZQ2 | Gene names | Afg3l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-). | |||||
|
YMEL1_MOUSE
|
||||||
| NC score | 0.943618 (rank : 20) | θ value | 3.72821e-46 (rank : 20) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
ATAD1_HUMAN
|
||||||
| NC score | 0.940978 (rank : 21) | θ value | 1.46948e-34 (rank : 36) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8NBU5, Q6P4B9, Q8N3G1, Q8WYR9, Q969Y3 | Gene names | ATAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
ATAD1_MOUSE
|
||||||
| NC score | 0.940978 (rank : 22) | θ value | 1.46948e-34 (rank : 37) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9D5T0, Q3U8V2, Q9D7A4 | Gene names | Atad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
NVL_MOUSE
|
||||||
| NC score | 0.940233 (rank : 23) | θ value | 1.05066e-40 (rank : 30) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
YMEL1_HUMAN
|
||||||
| NC score | 0.940047 (rank : 24) | θ value | 6.35935e-46 (rank : 21) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96TA2, Q9H1Q0, Q9UMR9 | Gene names | YME1L1, FTSH1, YME1L | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin- associated metalloprotease) (PAMP). | |||||
|
NVL_HUMAN
|
||||||
| NC score | 0.939766 (rank : 25) | θ value | 3.61106e-41 (rank : 29) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PEX6_HUMAN
|
||||||
| NC score | 0.938358 (rank : 26) | θ value | 2.18568e-46 (rank : 19) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PEX6_MOUSE
|
||||||
| NC score | 0.938122 (rank : 27) | θ value | 3.15612e-45 (rank : 22) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
KATL1_HUMAN
|
||||||
| NC score | 0.930187 (rank : 28) | θ value | 3.61106e-41 (rank : 28) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9BW62 | Gene names | KATNAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
KATL1_MOUSE
|
||||||
| NC score | 0.929876 (rank : 29) | θ value | 1.51715e-39 (rank : 31) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8K0T4 | Gene names | Katnal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
KTNA1_HUMAN
|
||||||
| NC score | 0.927278 (rank : 30) | θ value | 1.2411e-41 (rank : 26) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O75449, Q5TFA8, Q5TFA9, Q86VN2, Q9NU52 | Gene names | KATNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin). | |||||
|
NSF_MOUSE
|
||||||
| NC score | 0.926847 (rank : 31) | θ value | 3.4976e-36 (rank : 33) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P46460 | Gene names | Nsf, Skd2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein) (SKD2 protein). | |||||
|
NSF_HUMAN
|
||||||
| NC score | 0.926538 (rank : 32) | θ value | 2.05049e-36 (rank : 32) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P46459, Q9UKZ2 | Gene names | NSF | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein). | |||||
|
KTNA1_MOUSE
|
||||||
| NC score | 0.925077 (rank : 33) | θ value | 2.76489e-41 (rank : 27) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
|
SPAST_HUMAN
|
||||||
| NC score | 0.920727 (rank : 34) | θ value | 1.12514e-34 (rank : 34) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UBP0, Q9UPR9 | Gene names | SPAST, KIAA1083, SPG4 | |||
|
Domain Architecture |

|
|||||
| Description | Spastin. | |||||
|
SPAST_MOUSE
|
||||||
| NC score | 0.919217 (rank : 35) | θ value | 1.12514e-34 (rank : 35) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
VPS4B_HUMAN
|
||||||
| NC score | 0.911554 (rank : 36) | θ value | 4.00176e-32 (rank : 40) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4B_MOUSE
|
||||||
| NC score | 0.911029 (rank : 37) | θ value | 1.52067e-31 (rank : 41) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4A_HUMAN
|
||||||
| NC score | 0.909272 (rank : 38) | θ value | 2.77131e-33 (rank : 38) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| NC score | 0.909253 (rank : 39) | θ value | 2.77131e-33 (rank : 39) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
ATAD2_MOUSE
|
||||||
| NC score | 0.896276 (rank : 40) | θ value | 1.73182e-43 (rank : 25) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
ATAD2_HUMAN
|
||||||
| NC score | 0.879506 (rank : 41) | θ value | 2.67181e-44 (rank : 24) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
FIGN_MOUSE
|
||||||
| NC score | 0.874216 (rank : 42) | θ value | 3.87602e-27 (rank : 42) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
FIGN_HUMAN
|
||||||
| NC score | 0.868451 (rank : 43) | θ value | 1.47289e-26 (rank : 43) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
BCS1_HUMAN
|
||||||
| NC score | 0.718335 (rank : 44) | θ value | 4.59992e-12 (rank : 45) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y276, Q7Z2V7 | Gene names | BCS1L, BCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein) (H-BCS1). | |||||
|
BCS1_MOUSE
|
||||||
| NC score | 0.710968 (rank : 45) | θ value | 1.2105e-12 (rank : 44) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9CZP5 | Gene names | Bcs1l | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein). | |||||
|
TRP13_MOUSE
|
||||||
| NC score | 0.710382 (rank : 46) | θ value | 2.36244e-08 (rank : 49) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q3UA06, Q3UQG6, Q9CWW8 | Gene names | Trip13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13). | |||||
|
TRP13_HUMAN
|
||||||
| NC score | 0.706895 (rank : 47) | θ value | 5.26297e-08 (rank : 50) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q15645, O15324 | Gene names | TRIP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13) (Human papillomavirus type 16 E1 protein- binding protein) (HPV16 E1 protein-binding protein) (16E1-BP). | |||||
|
ATD3A_HUMAN
|
||||||
| NC score | 0.637878 (rank : 48) | θ value | 2.36244e-08 (rank : 48) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9NVI7, Q8N275, Q96A50 | Gene names | ATAD3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3A. | |||||
|
ATD3B_HUMAN
|
||||||
| NC score | 0.629192 (rank : 49) | θ value | 4.30538e-10 (rank : 46) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
ATAD3_MOUSE
|
||||||
| NC score | 0.619934 (rank : 50) | θ value | 2.79066e-09 (rank : 47) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
CLPX_HUMAN
|
||||||
| NC score | 0.459857 (rank : 51) | θ value | 1.87187e-05 (rank : 51) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
CLPX_MOUSE
|
||||||
| NC score | 0.457928 (rank : 52) | θ value | 1.87187e-05 (rank : 52) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JHS4, Q9WVD1 | Gene names | Clpx | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
LONM_HUMAN
|
||||||
| NC score | 0.440437 (rank : 53) | θ value | 0.00665767 (rank : 53) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
RUVB1_HUMAN
|
||||||
| NC score | 0.318985 (rank : 54) | θ value | 0.125558 (rank : 57) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y265, Q9BSX9 | Gene names | RUVBL1, NMP238, TIP49, TIP49A | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (Nuclear matrix protein 238) (NMP 238) (54 kDa erythrocyte cytosolic protein) (ECP-54) (TIP60-associated protein 54-alpha) (TAP54-alpha). | |||||
|
RUVB1_MOUSE
|
||||||
| NC score | 0.318947 (rank : 55) | θ value | 0.125558 (rank : 58) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P60122, O35753 | Gene names | Ruvbl1, Tip49, Tip49a | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (DNA helicase p50). | |||||
|
WRIP1_MOUSE
|
||||||
| NC score | 0.235981 (rank : 56) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
WRIP1_HUMAN
|
||||||
| NC score | 0.232252 (rank : 57) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
RFC1_MOUSE
|
||||||
| NC score | 0.130389 (rank : 58) | θ value | 0.163984 (rank : 59) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
RUVB2_HUMAN
|
||||||
| NC score | 0.126406 (rank : 59) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y230, Q6PK27, Q9Y361 | Gene names | RUVBL2, TIP48, TIP49B | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (48 kDa TATA box-binding protein-interacting protein) (48 kDa TBP-interacting protein) (TIP49b) (Repressing pontin 52) (Reptin 52) (51 kDa erythrocyte cytosolic protein) (ECP-51) (TIP60-associated protein 54-beta) (TAP54-beta). | |||||
|
RUVB2_MOUSE
|
||||||
| NC score | 0.123826 (rank : 60) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9WTM5 | Gene names | Ruvbl2 | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (p47 protein). | |||||
|
THNSL_MOUSE
|
||||||
| NC score | 0.118146 (rank : 61) | θ value | 1.38821 (rank : 78) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BH55 | Gene names | Thnsl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
THNSL_HUMAN
|
||||||
| NC score | 0.115815 (rank : 62) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IYQ7, Q5VV21 | Gene names | THNSL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
RFC1_HUMAN
|
||||||
| NC score | 0.115106 (rank : 63) | θ value | 0.62314 (rank : 62) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
RFC4_HUMAN
|
||||||
| NC score | 0.095602 (rank : 64) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P35249, Q6FHX7 | Gene names | RFC4 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 4 (Replication factor C 37 kDa subunit) (RF-C 37 kDa subunit) (RFC37) (Activator 1 37 kDa subunit) (A1 37 kDa subunit). | |||||
|
KAD6_HUMAN
|
||||||
| NC score | 0.090955 (rank : 65) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
RFC4_MOUSE
|
||||||
| NC score | 0.084139 (rank : 66) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99J62 | Gene names | Rfc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Replication factor C subunit 4. | |||||
|
KAD6_MOUSE
|
||||||
| NC score | 0.069932 (rank : 67) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VCP8 | Gene names | Ak6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
RFC5_MOUSE
|
||||||
| NC score | 0.065920 (rank : 68) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9D0F6 | Gene names | Rfc5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RFC5_HUMAN
|
||||||
| NC score | 0.064481 (rank : 69) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P40937 | Gene names | RFC5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RENT1_MOUSE
|
||||||
| NC score | 0.045190 (rank : 70) | θ value | 0.0961366 (rank : 54) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9EPU0, Q99PR4 | Gene names | Upf1, Rent1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (mUpf1). | |||||
|
RENT1_HUMAN
|
||||||
| NC score | 0.043742 (rank : 71) | θ value | 0.125558 (rank : 56) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92900, O00239, O43343, Q86Z25, Q92842 | Gene names | UPF1, KIAA0221, RENT1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (hUpf1). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.043216 (rank : 72) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
K2C4_HUMAN
|
||||||
| NC score | 0.031551 (rank : 73) | θ value | 0.21417 (rank : 60) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P19013, Q6GTR8, Q96LA7, Q9BTL1 | Gene names | KRT4, CYK4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 4 (Cytokeratin-4) (CK-4) (Keratin-4) (K4). | |||||
|
MOV10_HUMAN
|
||||||
| NC score | 0.030479 (rank : 74) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HCE1, Q8TEF0, Q9BSY3, Q9BUJ9 | Gene names | MOV10, KIAA1631 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase MOV-10 (EC 3.6.1.-) (Moloney leukemia virus 10 protein). | |||||
|
KRHB4_HUMAN
|
||||||
| NC score | 0.029890 (rank : 75) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NSB2 | Gene names | KRTHB4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb4 (Hair keratin, type II Hb4). | |||||
|
K2C1_MOUSE
|
||||||
| NC score | 0.029889 (rank : 76) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P04104 | Gene names | Krt1, Krt2-1 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin). | |||||
|
KRHB5_HUMAN
|
||||||
| NC score | 0.029305 (rank : 77) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P78386, Q9NSB1 | Gene names | KRTHB5 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb5 (Hair keratin, type II Hb5). | |||||
|
KRHB5_MOUSE
|
||||||
| NC score | 0.029287 (rank : 78) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Z2T6, Q8CE83, Q9D7M4 | Gene names | Krthb5, Krt2-18 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb5 (Hair keratin, type II Hb5). | |||||
|
KRHB3_HUMAN
|
||||||
| NC score | 0.029173 (rank : 79) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P78385, Q9NSB3 | Gene names | KRTHB3 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb3 (Hair keratin, type II Hb3). | |||||
|
KRHB1_HUMAN
|
||||||
| NC score | 0.029038 (rank : 80) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14533, Q14846, Q16274, Q8WU52, Q9BR74 | Gene names | KRTHB1, MLN137 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb1 (Hair keratin, type II Hb1) (ghHKb1) (ghHb1) (MLN 137). | |||||
|
KRHB6_HUMAN
|
||||||
| NC score | 0.029026 (rank : 81) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43790, P78387 | Gene names | KRTHB6 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb6 (Hair keratin, type II Hb6) (ghHb6). | |||||
|
K2C1B_MOUSE
|
||||||
| NC score | 0.028934 (rank : 82) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6IFZ6 | Gene names | Krt1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 1b (Type II keratin Kb39) (Embryonic type II keratin-1). | |||||
|
K22E_HUMAN
|
||||||
| NC score | 0.028855 (rank : 83) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 515 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35908 | Gene names | KRT2A, KRT2E | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 2 epidermal (Cytokeratin-2e) (K2e) (CK 2e). | |||||
|
K2C6A_MOUSE
|
||||||
| NC score | 0.028724 (rank : 84) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P50446, Q9Z332 | Gene names | Krt6a, Ker2, Krt2-6, Krt2-6a, Krt6 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6A (Cytokeratin-6A) (CK 6A) (K6a keratin) (Keratin-6 alpha) (mK6-alpha). | |||||
|
KRHB6_MOUSE
|
||||||
| NC score | 0.028652 (rank : 85) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97861, Q3TTV9 | Gene names | Krthb6, Krt2-10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cuticular Hb6 (Hair keratin, type II Hb6). | |||||
|
K2C6B_MOUSE
|
||||||
| NC score | 0.028627 (rank : 86) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Z331 | Gene names | Krt6b, K6-beta, Krt2-6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 6B (Cytokeratin-6B) (CK 6B) (K6b keratin) (Keratin-6 beta) (mK6-beta). | |||||
|
K2C6A_HUMAN
|
||||||
| NC score | 0.028598 (rank : 87) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P02538, Q6NT67, Q96CL4 | Gene names | KRT6A, K6A | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6A (Cytokeratin-6A) (CK 6A) (K6a keratin). | |||||
|
K2C6B_HUMAN
|
||||||
| NC score | 0.028558 (rank : 88) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P04259, P48669 | Gene names | KRT6B, K6B | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6B (Cytokeratin-6B) (CK 6B) (K6b keratin). | |||||
|
K2C6C_HUMAN
|
||||||
| NC score | 0.028538 (rank : 89) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P48666 | Gene names | KRT6C | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6C (Cytokeratin-6C) (CK 6C) (K6c keratin). | |||||
|
K2C6E_HUMAN
|
||||||
| NC score | 0.028509 (rank : 90) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P48668, Q7RTN9 | Gene names | KRT6E | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6E (Cytokeratin-6E) (CK 6E) (K6e keratin) (Keratin K6h). | |||||
|
K2C4_MOUSE
|
||||||
| NC score | 0.028371 (rank : 91) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P07744, Q6P3F5 | Gene names | Krt4, Krt2-4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 4 (Cytokeratin-4) (CK-4) (Keratin-4) (K4) (Cytoskeletal 57 kDa keratin). | |||||
|
KRHB4_MOUSE
|
||||||
| NC score | 0.028131 (rank : 92) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99M73 | Gene names | Krthb4, Krt2-16 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb4 (Hair keratin, type II Hb4) (65 kDa type II keratin). | |||||
|
K22O_HUMAN
|
||||||
| NC score | 0.028121 (rank : 93) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q01546, Q7Z795 | Gene names | KRT76, KRT2B, KRT2P | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 2 oral (Cytokeratin-2P) (K2P) (CK 2P) (Keratin 76). | |||||
|
K2C1_HUMAN
|
||||||
| NC score | 0.027957 (rank : 94) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P04264, Q14720, Q6GSJ0, Q9H298 | Gene names | KRT1, KRTA | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin) (Hair alpha protein). | |||||
|
K2C5_MOUSE
|
||||||
| NC score | 0.027945 (rank : 95) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q922U2, Q920F2 | Gene names | Krt5, Krt2-5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 5 (Cytokeratin-5) (CK-5) (Keratin-5) (K5). | |||||
|
K2C5_HUMAN
|
||||||
| NC score | 0.027853 (rank : 96) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P13647, Q6PI71, Q6UBJ0, Q8TA91 | Gene names | KRT5 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 5 (Cytokeratin-5) (CK-5) (Keratin-5) (K5) (58 kDa cytokeratin). | |||||
|
K2C8_MOUSE
|
||||||
| NC score | 0.027670 (rank : 97) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 576 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P11679, Q3KQK5, Q3TGI1, Q3TJE1, Q3TKY7, Q61463, Q61518, Q61519 | Gene names | Krt8, Krt2-8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8) (Cytokeratin endo A). | |||||
|
K2C8_HUMAN
|
||||||
| NC score | 0.026566 (rank : 98) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P05787, Q14099, Q14716, Q14717, Q53GJ0, Q6DHW5, Q6GMY0, Q96J60 | Gene names | KRT8, CYK8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8). | |||||
|
MOV10_MOUSE
|
||||||
| NC score | 0.025986 (rank : 99) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P23249, Q9DC64 | Gene names | Mov10, Gb110 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase MOV-10 (EC 3.6.1.-) (Moloney leukemia virus 10 protein). | |||||
|
M10L1_MOUSE
|
||||||
| NC score | 0.024025 (rank : 100) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99MV5, Q7TPA9, Q8C3W0, Q924C2 | Gene names | Mov10l1, Champ | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase Mov10l1 (EC 3.6.1.-) (Moloney leukemia virus 10-like protein 1) (MOV10-like 1) (Cardiac helicase activated by MEF2 protein) (Cardiac-specific RNA helicase). | |||||
|
TXLNG_MOUSE
|
||||||
| NC score | 0.018432 (rank : 101) | θ value | 1.81305 (rank : 80) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BHN1, Q8BP11 | Gene names | Txlng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin. | |||||
|
TXLNG_HUMAN
|
||||||
| NC score | 0.016718 (rank : 102) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NUQ3, Q9P0X1 | Gene names | TXLNG, CXorf15, LSR5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin (Lipopolysaccharide-specific response protein 5). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.013726 (rank : 103) | θ value | 0.365318 (rank : 61) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
TBCD4_HUMAN
|
||||||
| NC score | 0.013662 (rank : 104) | θ value | 0.813845 (rank : 63) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60343, Q5W0B9 | Gene names | TBC1D4, KIAA0603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 4 (Akt substrate of 160 kDa) (AS160). | |||||
|
RT15_MOUSE
|
||||||
| NC score | 0.013125 (rank : 105) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DC71, Q7TMG9, Q8C7F1, Q9CVE0, Q9CWX1 | Gene names | Mrps15, Rpms15 | |||
|
Domain Architecture |

|
|||||
| Description | 28S ribosomal protein S15, mitochondrial precursor (S15mt) (MRP-S15). | |||||
|
CR034_HUMAN
|
||||||
| NC score | 0.011148 (rank : 106) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5BJE1, Q6ZP67, Q6ZU20 | Gene names | C18orf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34. | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.011075 (rank : 107) | θ value | 0.125558 (rank : 55) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.011068 (rank : 108) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.009311 (rank : 109) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
RBP22_HUMAN
|
||||||
| NC score | 0.008428 (rank : 110) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q53T03, Q53QN2, Q59GM7 | Gene names | RANBP2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 2 (RanBP2L2). | |||||
|
IMA7_MOUSE
|
||||||
| NC score | 0.008086 (rank : 111) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35345, Q9CVP9 | Gene names | Kpna6, Kpna5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6 subunit) (Importin alpha S2). | |||||
|
PIBF1_HUMAN
|
||||||
| NC score | 0.008013 (rank : 112) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
NUP88_MOUSE
|
||||||
| NC score | 0.007442 (rank : 113) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CEC0, Q80Z13, Q8K090 | Gene names | Nup88 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore complex protein Nup88 (Nucleoporin Nup88) (88 kDa nuclear pore complex protein). | |||||
|
COG3_HUMAN
|
||||||
| NC score | 0.007143 (rank : 114) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JB2, Q5VT70, Q8IXX4, Q9BZ92 | Gene names | COG3, SEC34 | |||
|
Domain Architecture |

|
|||||
| Description | Conserved oligomeric Golgi complex component 3 (Vesicle docking protein SEC34 homolog) (p94). | |||||
|
HOOK3_MOUSE
|
||||||
| NC score | 0.006919 (rank : 115) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
BICD1_MOUSE
|
||||||
| NC score | 0.006903 (rank : 116) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
GCP6_HUMAN
|
||||||
| NC score | 0.006850 (rank : 117) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96RT7, Q5JZ80, Q6PJ40, Q86YE9, Q9BY91, Q9UGX3, Q9UGX4 | Gene names | TUBGCP6, GCP6, KIAA1669 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-tubulin complex component 6 (GCP-6). | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.006705 (rank : 118) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
RGPD8_HUMAN
|
||||||
| NC score | 0.005719 (rank : 119) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99666 | Gene names | RGPD8, RANBP2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RANBP2-like and GRIP domain-containing protein 8 (Ran-binding protein 2-like 1) (RanBP2L1) (Sperm membrane protein BS-63). | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.005543 (rank : 120) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.005187 (rank : 121) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.005124 (rank : 122) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
IQGA3_HUMAN
|
||||||
| NC score | 0.004419 (rank : 123) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86VI3 | Gene names | IQGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP3. | |||||