Please be patient as the page loads
|
PRS6A_MOUSE
|
||||||
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PRS6A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999752 (rank : 2) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P17980, Q96HD3 | Gene names | PSMC3, TBP1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1) (Proteasome subunit P50). | |||||
|
PRS6A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
|
PRS4_HUMAN
|
||||||
| θ value | 4.34313e-95 (rank : 3) | NC score | 0.988567 (rank : 3) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P62191, P49014, Q03527, Q6IAW0, Q96AZ3 | Gene names | PSMC1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS4_MOUSE
|
||||||
| θ value | 4.34313e-95 (rank : 4) | NC score | 0.988567 (rank : 4) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P62192, P49014, Q03527, Q96AZ3 | Gene names | Psmc1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS8_HUMAN
|
||||||
| θ value | 1.40038e-85 (rank : 5) | NC score | 0.985299 (rank : 5) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P62195, O35051, O43208, P47210, P52915, P52916 | Gene names | PSMC5, SUG1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (Thyroid hormone receptor- interacting protein 1) (TRIP1). | |||||
|
PRS8_MOUSE
|
||||||
| θ value | 1.40038e-85 (rank : 6) | NC score | 0.985299 (rank : 6) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P62196, O35051, P47210, P52915, P52916, Q3UL51, Q9CWN5 | Gene names | Psmc5, Sug1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (mSUG1). | |||||
|
PRS6B_HUMAN
|
||||||
| θ value | 3.11973e-85 (rank : 7) | NC score | 0.985007 (rank : 7) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P43686, Q96FV5, Q9UBM3, Q9UEX3 | Gene names | PSMC4, TBP7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7). | |||||
|
PRS6B_MOUSE
|
||||||
| θ value | 3.44927e-84 (rank : 8) | NC score | 0.984601 (rank : 10) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P54775 | Gene names | Psmc4, Tbp7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7) (CIP21). | |||||
|
PRS7_HUMAN
|
||||||
| θ value | 3.34088e-79 (rank : 9) | NC score | 0.984625 (rank : 8) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P35998 | Gene names | PSMC2, MSS1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS7_MOUSE
|
||||||
| θ value | 3.34088e-79 (rank : 10) | NC score | 0.984614 (rank : 9) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P46471, O08531, Q3TGX5, Q9DBA1 | Gene names | Psmc2, Mss1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS10_HUMAN
|
||||||
| θ value | 2.92676e-75 (rank : 11) | NC score | 0.983995 (rank : 11) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P62333, P49719, Q6IBU3, Q92524 | Gene names | PSMC6, SUG2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS10_MOUSE
|
||||||
| θ value | 2.92676e-75 (rank : 12) | NC score | 0.983995 (rank : 12) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P62334, P49719, Q3TKK1, Q810A6, Q92524, Q9CXH9 | Gene names | Psmc6, Sug2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
TERA_HUMAN
|
||||||
| θ value | 7.25919e-50 (rank : 13) | NC score | 0.949823 (rank : 13) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P55072, Q969G7 | Gene names | VCP | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
TERA_MOUSE
|
||||||
| θ value | 9.48078e-50 (rank : 14) | NC score | 0.949770 (rank : 14) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q01853 | Gene names | Vcp | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
AFG32_MOUSE
|
||||||
| θ value | 8.87376e-48 (rank : 15) | NC score | 0.939953 (rank : 21) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8JZQ2 | Gene names | Afg3l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-). | |||||
|
AFG32_HUMAN
|
||||||
| θ value | 1.97686e-47 (rank : 16) | NC score | 0.940078 (rank : 20) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y4W6 | Gene names | AFG3L2 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-) (Paraplegin-like protein). | |||||
|
AFG31_MOUSE
|
||||||
| θ value | 3.72821e-46 (rank : 17) | NC score | 0.943256 (rank : 17) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q920A7 | Gene names | Afg3l1 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 1 (EC 3.4.24.-). | |||||
|
NVL_MOUSE
|
||||||
| θ value | 1.32601e-43 (rank : 18) | NC score | 0.938539 (rank : 22) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
NVL_HUMAN
|
||||||
| θ value | 1.12253e-42 (rank : 19) | NC score | 0.937692 (rank : 23) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
ATAD2_HUMAN
|
||||||
| θ value | 7.27602e-42 (rank : 20) | NC score | 0.878294 (rank : 41) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
PEX6_HUMAN
|
||||||
| θ value | 2.76489e-41 (rank : 21) | NC score | 0.933342 (rank : 25) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
ATAD2_MOUSE
|
||||||
| θ value | 1.3722e-40 (rank : 22) | NC score | 0.894334 (rank : 40) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
SPG7_HUMAN
|
||||||
| θ value | 1.3722e-40 (rank : 23) | NC score | 0.944940 (rank : 16) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UQ90, O75756, Q96IB0 | Gene names | SPG7, CAR, CMAR, PGN | |||
|
Domain Architecture |

|
|||||
| Description | Paraplegin (EC 3.4.24.-) (Spastic paraplegia protein 7). | |||||
|
PEX1_HUMAN
|
||||||
| θ value | 1.79215e-40 (rank : 24) | NC score | 0.945782 (rank : 15) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
PEX6_MOUSE
|
||||||
| θ value | 5.21438e-40 (rank : 25) | NC score | 0.932871 (rank : 26) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
YMEL1_HUMAN
|
||||||
| θ value | 3.37985e-39 (rank : 26) | NC score | 0.932089 (rank : 27) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96TA2, Q9H1Q0, Q9UMR9 | Gene names | YME1L1, FTSH1, YME1L | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin- associated metalloprotease) (PAMP). | |||||
|
YMEL1_MOUSE
|
||||||
| θ value | 3.37985e-39 (rank : 27) | NC score | 0.935608 (rank : 24) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
KATL1_MOUSE
|
||||||
| θ value | 2.19075e-38 (rank : 28) | NC score | 0.927380 (rank : 29) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8K0T4 | Gene names | Katnal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
KATL1_HUMAN
|
||||||
| θ value | 4.88048e-38 (rank : 29) | NC score | 0.927387 (rank : 28) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9BW62 | Gene names | KATNAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
KTNA1_MOUSE
|
||||||
| θ value | 8.32485e-38 (rank : 30) | NC score | 0.922854 (rank : 31) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
|
KTNA1_HUMAN
|
||||||
| θ value | 2.42216e-37 (rank : 31) | NC score | 0.924634 (rank : 30) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O75449, Q5TFA8, Q5TFA9, Q86VN2, Q9NU52 | Gene names | KATNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin). | |||||
|
ATAD1_HUMAN
|
||||||
| θ value | 3.86705e-35 (rank : 32) | NC score | 0.940268 (rank : 18) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8NBU5, Q6P4B9, Q8N3G1, Q8WYR9, Q969Y3 | Gene names | ATAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
ATAD1_MOUSE
|
||||||
| θ value | 3.86705e-35 (rank : 33) | NC score | 0.940268 (rank : 19) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9D5T0, Q3U8V2, Q9D7A4 | Gene names | Atad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
SPAST_HUMAN
|
||||||
| θ value | 1.6247e-33 (rank : 34) | NC score | 0.919169 (rank : 33) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UBP0, Q9UPR9 | Gene names | SPAST, KIAA1083, SPG4 | |||
|
Domain Architecture |

|
|||||
| Description | Spastin. | |||||
|
VPS4B_MOUSE
|
||||||
| θ value | 2.77131e-33 (rank : 35) | NC score | 0.910401 (rank : 37) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
SPAST_MOUSE
|
||||||
| θ value | 4.72714e-33 (rank : 36) | NC score | 0.917412 (rank : 35) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
VPS4B_HUMAN
|
||||||
| θ value | 4.72714e-33 (rank : 37) | NC score | 0.910656 (rank : 36) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4A_HUMAN
|
||||||
| θ value | 1.98606e-31 (rank : 38) | NC score | 0.907390 (rank : 38) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| θ value | 1.98606e-31 (rank : 39) | NC score | 0.907369 (rank : 39) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
NSF_MOUSE
|
||||||
| θ value | 2.59387e-31 (rank : 40) | NC score | 0.919674 (rank : 32) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P46460 | Gene names | Nsf, Skd2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein) (SKD2 protein). | |||||
|
NSF_HUMAN
|
||||||
| θ value | 4.42448e-31 (rank : 41) | NC score | 0.919118 (rank : 34) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P46459, Q9UKZ2 | Gene names | NSF | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein). | |||||
|
FIGN_MOUSE
|
||||||
| θ value | 4.73814e-25 (rank : 42) | NC score | 0.873859 (rank : 42) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
FIGN_HUMAN
|
||||||
| θ value | 8.08199e-25 (rank : 43) | NC score | 0.868258 (rank : 43) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
ATAD3_MOUSE
|
||||||
| θ value | 9.59137e-10 (rank : 44) | NC score | 0.624299 (rank : 50) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
ATD3B_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 45) | NC score | 0.631098 (rank : 49) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
ATD3A_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 46) | NC score | 0.642324 (rank : 48) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9NVI7, Q8N275, Q96A50 | Gene names | ATAD3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3A. | |||||
|
BCS1_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 47) | NC score | 0.699657 (rank : 44) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y276, Q7Z2V7 | Gene names | BCS1L, BCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein) (H-BCS1). | |||||
|
BCS1_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 48) | NC score | 0.690644 (rank : 45) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9CZP5 | Gene names | Bcs1l | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein). | |||||
|
CLPX_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 49) | NC score | 0.449977 (rank : 52) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JHS4, Q9WVD1 | Gene names | Clpx | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
CLPX_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 50) | NC score | 0.451823 (rank : 51) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
TRP13_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 51) | NC score | 0.683967 (rank : 46) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q3UA06, Q3UQG6, Q9CWW8 | Gene names | Trip13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13). | |||||
|
TRP13_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 52) | NC score | 0.680171 (rank : 47) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q15645, O15324 | Gene names | TRIP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13) (Human papillomavirus type 16 E1 protein- binding protein) (HPV16 E1 protein-binding protein) (16E1-BP). | |||||
|
RFC5_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 53) | NC score | 0.091967 (rank : 66) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9D0F6 | Gene names | Rfc5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RFC5_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 54) | NC score | 0.090599 (rank : 68) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P40937 | Gene names | RFC5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
WRIP1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 55) | NC score | 0.249560 (rank : 57) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
RUVB1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 56) | NC score | 0.308706 (rank : 54) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y265, Q9BSX9 | Gene names | RUVBL1, NMP238, TIP49, TIP49A | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (Nuclear matrix protein 238) (NMP 238) (54 kDa erythrocyte cytosolic protein) (ECP-54) (TIP60-associated protein 54-alpha) (TAP54-alpha). | |||||
|
RUVB1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 57) | NC score | 0.308667 (rank : 55) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P60122, O35753 | Gene names | Ruvbl1, Tip49, Tip49a | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (DNA helicase p50). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 58) | NC score | 0.014766 (rank : 76) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
WRIP1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 59) | NC score | 0.252233 (rank : 56) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
TAXB1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.010259 (rank : 80) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
LONM_HUMAN
|
||||||
| θ value | 0.813845 (rank : 61) | NC score | 0.419752 (rank : 53) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
SMC2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 62) | NC score | 0.013133 (rank : 77) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.012913 (rank : 78) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CCD13_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.006655 (rank : 83) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
NAB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.021343 (rank : 74) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13506, O75383, O75384, Q6GTU1, Q9UEV1 | Gene names | NAB1 | |||
|
Domain Architecture |
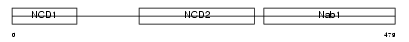
|
|||||
| Description | NGFI-A-binding protein 1 (EGR-1-binding protein 1) (Transcriptional regulatory protein p54). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.004827 (rank : 88) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
MYH14_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.001982 (rank : 92) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
NAB1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.018726 (rank : 75) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61122 | Gene names | Nab1 | |||
|
Domain Architecture |
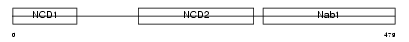
|
|||||
| Description | NGFI-A-binding protein 1 (EGR-1-binding protein 1). | |||||
|
RBP22_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.009059 (rank : 82) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q53T03, Q53QN2, Q59GM7 | Gene names | RANBP2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 2 (RanBP2L2). | |||||
|
SMC4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.009705 (rank : 81) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CG47, Q8BTS7, Q8BTY9, Q99K21 | Gene names | Smc4l1, Capc, Smc4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (XCAP-C homolog). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.006110 (rank : 85) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
DOCK8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.006415 (rank : 84) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NF50, Q3MV16, Q5JPJ1, Q8TEP1, Q8WUY2, Q9BYJ5, Q9H1Q2, Q9H1Q3, Q9H308, Q9H7P2 | Gene names | DOCK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dedicator of cytokinesis protein 8. | |||||
|
OPTN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.005750 (rank : 86) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
PEPL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.003053 (rank : 90) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
RFC4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.111798 (rank : 58) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P35249, Q6FHX7 | Gene names | RFC4 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 4 (Replication factor C 37 kDa subunit) (RF-C 37 kDa subunit) (RFC37) (Activator 1 37 kDa subunit) (A1 37 kDa subunit). | |||||
|
RFC4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.100818 (rank : 62) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99J62 | Gene names | Rfc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Replication factor C subunit 4. | |||||
|
SPAG5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.012032 (rank : 79) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TME2, Q8CH61 | Gene names | Spag5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Mastrin) (Mitotic spindle-associated protein p126) (MAP126). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.002291 (rank : 91) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
KINH_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.001843 (rank : 93) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
NAF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.003618 (rank : 89) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
RENT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.025028 (rank : 72) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9EPU0, Q99PR4 | Gene names | Upf1, Rent1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (mUpf1). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.036232 (rank : 71) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
MORC4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.005712 (rank : 87) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BMD7, Q4KMM6, Q8BX95, Q9CS96 | Gene names | Morc4, Zcwcc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
RENT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.023447 (rank : 73) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92900, O00239, O43343, Q86Z25, Q92842 | Gene names | UPF1, KIAA0221, RENT1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (hUpf1). | |||||
|
UACA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.000328 (rank : 94) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
KAD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.090932 (rank : 67) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
KAD6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.070425 (rank : 69) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VCP8 | Gene names | Ak6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
RFC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.098855 (rank : 64) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
RFC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.111189 (rank : 59) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
RFC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.050582 (rank : 70) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P35250, P32846, Q9BU93 | Gene names | RFC2 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 2 (Replication factor C 40 kDa subunit) (RF-C 40 kDa subunit) (RFC40) (Activator 1 40 kDa subunit) (A1 40 kDa subunit). | |||||
|
RUVB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.110868 (rank : 60) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y230, Q6PK27, Q9Y361 | Gene names | RUVBL2, TIP48, TIP49B | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (48 kDa TATA box-binding protein-interacting protein) (48 kDa TBP-interacting protein) (TIP49b) (Repressing pontin 52) (Reptin 52) (51 kDa erythrocyte cytosolic protein) (ECP-51) (TIP60-associated protein 54-beta) (TAP54-beta). | |||||
|
RUVB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.108271 (rank : 61) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9WTM5 | Gene names | Ruvbl2 | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (p47 protein). | |||||
|
THNSL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.095754 (rank : 65) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IYQ7, Q5VV21 | Gene names | THNSL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
THNSL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.099048 (rank : 63) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BH55 | Gene names | Thnsl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
PRS6A_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
|
PRS6A_HUMAN
|
||||||
| NC score | 0.999752 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P17980, Q96HD3 | Gene names | PSMC3, TBP1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1) (Proteasome subunit P50). | |||||
|
PRS4_HUMAN
|
||||||
| NC score | 0.988567 (rank : 3) | θ value | 4.34313e-95 (rank : 3) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P62191, P49014, Q03527, Q6IAW0, Q96AZ3 | Gene names | PSMC1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS4_MOUSE
|
||||||
| NC score | 0.988567 (rank : 4) | θ value | 4.34313e-95 (rank : 4) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P62192, P49014, Q03527, Q96AZ3 | Gene names | Psmc1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS8_HUMAN
|
||||||
| NC score | 0.985299 (rank : 5) | θ value | 1.40038e-85 (rank : 5) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P62195, O35051, O43208, P47210, P52915, P52916 | Gene names | PSMC5, SUG1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (Thyroid hormone receptor- interacting protein 1) (TRIP1). | |||||
|
PRS8_MOUSE
|
||||||
| NC score | 0.985299 (rank : 6) | θ value | 1.40038e-85 (rank : 6) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P62196, O35051, P47210, P52915, P52916, Q3UL51, Q9CWN5 | Gene names | Psmc5, Sug1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (mSUG1). | |||||
|
PRS6B_HUMAN
|
||||||
| NC score | 0.985007 (rank : 7) | θ value | 3.11973e-85 (rank : 7) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P43686, Q96FV5, Q9UBM3, Q9UEX3 | Gene names | PSMC4, TBP7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7). | |||||
|
PRS7_HUMAN
|
||||||
| NC score | 0.984625 (rank : 8) | θ value | 3.34088e-79 (rank : 9) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P35998 | Gene names | PSMC2, MSS1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS7_MOUSE
|
||||||
| NC score | 0.984614 (rank : 9) | θ value | 3.34088e-79 (rank : 10) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P46471, O08531, Q3TGX5, Q9DBA1 | Gene names | Psmc2, Mss1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS6B_MOUSE
|
||||||
| NC score | 0.984601 (rank : 10) | θ value | 3.44927e-84 (rank : 8) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P54775 | Gene names | Psmc4, Tbp7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7) (CIP21). | |||||
|
PRS10_HUMAN
|
||||||
| NC score | 0.983995 (rank : 11) | θ value | 2.92676e-75 (rank : 11) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P62333, P49719, Q6IBU3, Q92524 | Gene names | PSMC6, SUG2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS10_MOUSE
|
||||||
| NC score | 0.983995 (rank : 12) | θ value | 2.92676e-75 (rank : 12) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P62334, P49719, Q3TKK1, Q810A6, Q92524, Q9CXH9 | Gene names | Psmc6, Sug2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
TERA_HUMAN
|
||||||
| NC score | 0.949823 (rank : 13) | θ value | 7.25919e-50 (rank : 13) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P55072, Q969G7 | Gene names | VCP | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
TERA_MOUSE
|
||||||
| NC score | 0.949770 (rank : 14) | θ value | 9.48078e-50 (rank : 14) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q01853 | Gene names | Vcp | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
PEX1_HUMAN
|
||||||
| NC score | 0.945782 (rank : 15) | θ value | 1.79215e-40 (rank : 24) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
SPG7_HUMAN
|
||||||
| NC score | 0.944940 (rank : 16) | θ value | 1.3722e-40 (rank : 23) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UQ90, O75756, Q96IB0 | Gene names | SPG7, CAR, CMAR, PGN | |||
|
Domain Architecture |

|
|||||
| Description | Paraplegin (EC 3.4.24.-) (Spastic paraplegia protein 7). | |||||
|
AFG31_MOUSE
|
||||||
| NC score | 0.943256 (rank : 17) | θ value | 3.72821e-46 (rank : 17) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q920A7 | Gene names | Afg3l1 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 1 (EC 3.4.24.-). | |||||
|
ATAD1_HUMAN
|
||||||
| NC score | 0.940268 (rank : 18) | θ value | 3.86705e-35 (rank : 32) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8NBU5, Q6P4B9, Q8N3G1, Q8WYR9, Q969Y3 | Gene names | ATAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
ATAD1_MOUSE
|
||||||
| NC score | 0.940268 (rank : 19) | θ value | 3.86705e-35 (rank : 33) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9D5T0, Q3U8V2, Q9D7A4 | Gene names | Atad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
AFG32_HUMAN
|
||||||
| NC score | 0.940078 (rank : 20) | θ value | 1.97686e-47 (rank : 16) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y4W6 | Gene names | AFG3L2 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-) (Paraplegin-like protein). | |||||
|
AFG32_MOUSE
|
||||||
| NC score | 0.939953 (rank : 21) | θ value | 8.87376e-48 (rank : 15) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8JZQ2 | Gene names | Afg3l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-). | |||||
|
NVL_MOUSE
|
||||||
| NC score | 0.938539 (rank : 22) | θ value | 1.32601e-43 (rank : 18) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
NVL_HUMAN
|
||||||
| NC score | 0.937692 (rank : 23) | θ value | 1.12253e-42 (rank : 19) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
YMEL1_MOUSE
|
||||||
| NC score | 0.935608 (rank : 24) | θ value | 3.37985e-39 (rank : 27) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
PEX6_HUMAN
|
||||||
| NC score | 0.933342 (rank : 25) | θ value | 2.76489e-41 (rank : 21) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PEX6_MOUSE
|
||||||
| NC score | 0.932871 (rank : 26) | θ value | 5.21438e-40 (rank : 25) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
YMEL1_HUMAN
|
||||||
| NC score | 0.932089 (rank : 27) | θ value | 3.37985e-39 (rank : 26) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96TA2, Q9H1Q0, Q9UMR9 | Gene names | YME1L1, FTSH1, YME1L | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin- associated metalloprotease) (PAMP). | |||||
|
KATL1_HUMAN
|
||||||
| NC score | 0.927387 (rank : 28) | θ value | 4.88048e-38 (rank : 29) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9BW62 | Gene names | KATNAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
KATL1_MOUSE
|
||||||
| NC score | 0.927380 (rank : 29) | θ value | 2.19075e-38 (rank : 28) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8K0T4 | Gene names | Katnal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
KTNA1_HUMAN
|
||||||
| NC score | 0.924634 (rank : 30) | θ value | 2.42216e-37 (rank : 31) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O75449, Q5TFA8, Q5TFA9, Q86VN2, Q9NU52 | Gene names | KATNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin). | |||||
|
KTNA1_MOUSE
|
||||||
| NC score | 0.922854 (rank : 31) | θ value | 8.32485e-38 (rank : 30) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
|
NSF_MOUSE
|
||||||
| NC score | 0.919674 (rank : 32) | θ value | 2.59387e-31 (rank : 40) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P46460 | Gene names | Nsf, Skd2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein) (SKD2 protein). | |||||
|
SPAST_HUMAN
|
||||||
| NC score | 0.919169 (rank : 33) | θ value | 1.6247e-33 (rank : 34) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UBP0, Q9UPR9 | Gene names | SPAST, KIAA1083, SPG4 | |||
|
Domain Architecture |

|
|||||
| Description | Spastin. | |||||
|
NSF_HUMAN
|
||||||
| NC score | 0.919118 (rank : 34) | θ value | 4.42448e-31 (rank : 41) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P46459, Q9UKZ2 | Gene names | NSF | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein). | |||||
|
SPAST_MOUSE
|
||||||
| NC score | 0.917412 (rank : 35) | θ value | 4.72714e-33 (rank : 36) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
VPS4B_HUMAN
|
||||||
| NC score | 0.910656 (rank : 36) | θ value | 4.72714e-33 (rank : 37) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4B_MOUSE
|
||||||
| NC score | 0.910401 (rank : 37) | θ value | 2.77131e-33 (rank : 35) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4A_HUMAN
|
||||||
| NC score | 0.907390 (rank : 38) | θ value | 1.98606e-31 (rank : 38) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| NC score | 0.907369 (rank : 39) | θ value | 1.98606e-31 (rank : 39) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
ATAD2_MOUSE
|
||||||
| NC score | 0.894334 (rank : 40) | θ value | 1.3722e-40 (rank : 22) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
ATAD2_HUMAN
|
||||||
| NC score | 0.878294 (rank : 41) | θ value | 7.27602e-42 (rank : 20) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
FIGN_MOUSE
|
||||||
| NC score | 0.873859 (rank : 42) | θ value | 4.73814e-25 (rank : 42) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
FIGN_HUMAN
|
||||||
| NC score | 0.868258 (rank : 43) | θ value | 8.08199e-25 (rank : 43) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
BCS1_HUMAN
|
||||||
| NC score | 0.699657 (rank : 44) | θ value | 3.08544e-08 (rank : 47) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y276, Q7Z2V7 | Gene names | BCS1L, BCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein) (H-BCS1). | |||||
|
BCS1_MOUSE
|
||||||
| NC score | 0.690644 (rank : 45) | θ value | 6.87365e-08 (rank : 48) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9CZP5 | Gene names | Bcs1l | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein). | |||||
|
TRP13_MOUSE
|
||||||
| NC score | 0.683967 (rank : 46) | θ value | 0.000786445 (rank : 51) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q3UA06, Q3UQG6, Q9CWW8 | Gene names | Trip13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13). | |||||
|
TRP13_HUMAN
|
||||||
| NC score | 0.680171 (rank : 47) | θ value | 0.00102713 (rank : 52) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q15645, O15324 | Gene names | TRIP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13) (Human papillomavirus type 16 E1 protein- binding protein) (HPV16 E1 protein-binding protein) (16E1-BP). | |||||
|
ATD3A_HUMAN
|
||||||
| NC score | 0.642324 (rank : 48) | θ value | 3.64472e-09 (rank : 46) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9NVI7, Q8N275, Q96A50 | Gene names | ATAD3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3A. | |||||
|
ATD3B_HUMAN
|
||||||
| NC score | 0.631098 (rank : 49) | θ value | 1.63604e-09 (rank : 45) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
ATAD3_MOUSE
|
||||||
| NC score | 0.624299 (rank : 50) | θ value | 9.59137e-10 (rank : 44) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
CLPX_HUMAN
|
||||||
| NC score | 0.451823 (rank : 51) | θ value | 0.000270298 (rank : 50) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
CLPX_MOUSE
|
||||||
| NC score | 0.449977 (rank : 52) | θ value | 0.00020696 (rank : 49) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JHS4, Q9WVD1 | Gene names | Clpx | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
LONM_HUMAN
|
||||||
| NC score | 0.419752 (rank : 53) | θ value | 0.813845 (rank : 61) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
RUVB1_HUMAN
|
||||||
| NC score | 0.308706 (rank : 54) | θ value | 0.163984 (rank : 56) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y265, Q9BSX9 | Gene names | RUVBL1, NMP238, TIP49, TIP49A | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (Nuclear matrix protein 238) (NMP 238) (54 kDa erythrocyte cytosolic protein) (ECP-54) (TIP60-associated protein 54-alpha) (TAP54-alpha). | |||||
|
RUVB1_MOUSE
|
||||||
| NC score | 0.308667 (rank : 55) | θ value | 0.163984 (rank : 57) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P60122, O35753 | Gene names | Ruvbl1, Tip49, Tip49a | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (DNA helicase p50). | |||||
|
WRIP1_MOUSE
|
||||||
| NC score | 0.252233 (rank : 56) | θ value | 0.279714 (rank : 59) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
WRIP1_HUMAN
|
||||||
| NC score | 0.249560 (rank : 57) | θ value | 0.125558 (rank : 55) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
RFC4_HUMAN
|
||||||
| NC score | 0.111798 (rank : 58) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P35249, Q6FHX7 | Gene names | RFC4 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 4 (Replication factor C 37 kDa subunit) (RF-C 37 kDa subunit) (RFC37) (Activator 1 37 kDa subunit) (A1 37 kDa subunit). | |||||
|
RFC1_MOUSE
|
||||||
| NC score | 0.111189 (rank : 59) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
RUVB2_HUMAN
|
||||||
| NC score | 0.110868 (rank : 60) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y230, Q6PK27, Q9Y361 | Gene names | RUVBL2, TIP48, TIP49B | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (48 kDa TATA box-binding protein-interacting protein) (48 kDa TBP-interacting protein) (TIP49b) (Repressing pontin 52) (Reptin 52) (51 kDa erythrocyte cytosolic protein) (ECP-51) (TIP60-associated protein 54-beta) (TAP54-beta). | |||||
|
RUVB2_MOUSE
|
||||||
| NC score | 0.108271 (rank : 61) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9WTM5 | Gene names | Ruvbl2 | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (p47 protein). | |||||
|
RFC4_MOUSE
|
||||||
| NC score | 0.100818 (rank : 62) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99J62 | Gene names | Rfc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Replication factor C subunit 4. | |||||
|
THNSL_MOUSE
|
||||||
| NC score | 0.099048 (rank : 63) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BH55 | Gene names | Thnsl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
RFC1_HUMAN
|
||||||
| NC score | 0.098855 (rank : 64) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
THNSL_HUMAN
|
||||||
| NC score | 0.095754 (rank : 65) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IYQ7, Q5VV21 | Gene names | THNSL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
RFC5_MOUSE
|
||||||
| NC score | 0.091967 (rank : 66) | θ value | 0.0431538 (rank : 53) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9D0F6 | Gene names | Rfc5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
KAD6_HUMAN
|
||||||
| NC score | 0.090932 (rank : 67) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
RFC5_HUMAN
|
||||||
| NC score | 0.090599 (rank : 68) | θ value | 0.0563607 (rank : 54) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P40937 | Gene names | RFC5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
KAD6_MOUSE
|
||||||
| NC score | 0.070425 (rank : 69) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VCP8 | Gene names | Ak6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
RFC2_HUMAN
|
||||||
| NC score | 0.050582 (rank : 70) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P35250, P32846, Q9BU93 | Gene names | RFC2 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 2 (Replication factor C 40 kDa subunit) (RF-C 40 kDa subunit) (RFC40) (Activator 1 40 kDa subunit) (A1 40 kDa subunit). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.036232 (rank : 71) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
RENT1_MOUSE
|
||||||
| NC score | 0.025028 (rank : 72) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9EPU0, Q99PR4 | Gene names | Upf1, Rent1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (mUpf1). | |||||
|
RENT1_HUMAN
|
||||||
| NC score | 0.023447 (rank : 73) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92900, O00239, O43343, Q86Z25, Q92842 | Gene names | UPF1, KIAA0221, RENT1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (hUpf1). | |||||
|
NAB1_HUMAN
|
||||||
| NC score | 0.021343 (rank : 74) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13506, O75383, O75384, Q6GTU1, Q9UEV1 | Gene names | NAB1 | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 1 (EGR-1-binding protein 1) (Transcriptional regulatory protein p54). | |||||
|
NAB1_MOUSE
|
||||||
| NC score | 0.018726 (rank : 75) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61122 | Gene names | Nab1 | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 1 (EGR-1-binding protein 1). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.014766 (rank : 76) | θ value | 0.279714 (rank : 58) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
SMC2_MOUSE
|
||||||
| NC score | 0.013133 (rank : 77) | θ value | 0.813845 (rank : 62) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.012913 (rank : 78) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
SPAG5_MOUSE
|
||||||
| NC score | 0.012032 (rank : 79) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TME2, Q8CH61 | Gene names | Spag5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Mastrin) (Mitotic spindle-associated protein p126) (MAP126). | |||||
|
TAXB1_HUMAN
|
||||||
| NC score | 0.010259 (rank : 80) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
SMC4_MOUSE
|
||||||
| NC score | 0.009705 (rank : 81) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CG47, Q8BTS7, Q8BTY9, Q99K21 | Gene names | Smc4l1, Capc, Smc4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (XCAP-C homolog). | |||||
|
RBP22_HUMAN
|
||||||
| NC score | 0.009059 (rank : 82) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q53T03, Q53QN2, Q59GM7 | Gene names | RANBP2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 2 (RanBP2L2). | |||||
|
CCD13_HUMAN
|
||||||
| NC score | 0.006655 (rank : 83) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
DOCK8_HUMAN
|
||||||
| NC score | 0.006415 (rank : 84) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NF50, Q3MV16, Q5JPJ1, Q8TEP1, Q8WUY2, Q9BYJ5, Q9H1Q2, Q9H1Q3, Q9H308, Q9H7P2 | Gene names | DOCK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dedicator of cytokinesis protein 8. | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.006110 (rank : 85) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.005750 (rank : 86) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
MORC4_MOUSE
|
||||||
| NC score | 0.005712 (rank : 87) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BMD7, Q4KMM6, Q8BX95, Q9CS96 | Gene names | Morc4, Zcwcc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.004827 (rank : 88) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
NAF1_MOUSE
|
||||||
| NC score | 0.003618 (rank : 89) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.003053 (rank : 90) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.002291 (rank : 91) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.001982 (rank : 92) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
KINH_HUMAN
|
||||||
| NC score | 0.001843 (rank : 93) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
UACA_HUMAN
|
||||||
| NC score | 0.000328 (rank : 94) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||