Please be patient as the page loads
|
RFC2_HUMAN
|
||||||
| SwissProt Accessions | P35250, P32846, Q9BU93 | Gene names | RFC2 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 2 (Replication factor C 40 kDa subunit) (RF-C 40 kDa subunit) (RFC40) (Activator 1 40 kDa subunit) (A1 40 kDa subunit). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RFC2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35250, P32846, Q9BU93 | Gene names | RFC2 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 2 (Replication factor C 40 kDa subunit) (RF-C 40 kDa subunit) (RFC40) (Activator 1 40 kDa subunit) (A1 40 kDa subunit). | |||||
|
RFC2_MOUSE
|
||||||
| θ value | 6.5317e-184 (rank : 2) | NC score | 0.999332 (rank : 2) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9WUK4 | Gene names | Rfc2 | |||
|
Domain Architecture |
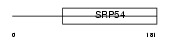
|
|||||
| Description | Replication factor C subunit 2 (Replication factor C 40 kDa subunit) (RF-C 40 kDa subunit) (RFC40) (Activator 1 40 kDa subunit) (A1 40 kDa subunit). | |||||
|
RFC5_HUMAN
|
||||||
| θ value | 1.0458e-56 (rank : 3) | NC score | 0.928570 (rank : 3) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P40937 | Gene names | RFC5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RFC5_MOUSE
|
||||||
| θ value | 1.97229e-55 (rank : 4) | NC score | 0.928150 (rank : 4) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D0F6 | Gene names | Rfc5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RFC4_HUMAN
|
||||||
| θ value | 8.57503e-51 (rank : 5) | NC score | 0.912155 (rank : 5) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P35249, Q6FHX7 | Gene names | RFC4 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 4 (Replication factor C 37 kDa subunit) (RF-C 37 kDa subunit) (RFC37) (Activator 1 37 kDa subunit) (A1 37 kDa subunit). | |||||
|
RFC4_MOUSE
|
||||||
| θ value | 3.04986e-48 (rank : 6) | NC score | 0.909806 (rank : 6) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99J62 | Gene names | Rfc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Replication factor C subunit 4. | |||||
|
RFC3_HUMAN
|
||||||
| θ value | 6.40375e-22 (rank : 7) | NC score | 0.770951 (rank : 8) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P40938, O15252 | Gene names | RFC3 | |||
|
Domain Architecture |
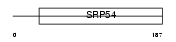
|
|||||
| Description | Replication factor C subunit 3 (Replication factor C 38 kDa subunit) (RFC38) (Activator 1 38 kDa subunit) (A1 38 kDa subunit) (RF-C 38 kDa subunit). | |||||
|
RFC3_MOUSE
|
||||||
| θ value | 6.40375e-22 (rank : 8) | NC score | 0.776148 (rank : 7) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R323 | Gene names | Rfc3 | |||
|
Domain Architecture |
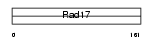
|
|||||
| Description | Replication factor C subunit 3 (Replication factor C 38 kDa subunit) (RFC38) (Activator 1 38 kDa subunit) (A1 38 kDa subunit) (RF-C 38 kDa subunit). | |||||
|
RFC1_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 9) | NC score | 0.412339 (rank : 9) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
RFC1_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 10) | NC score | 0.398793 (rank : 11) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
WRIP1_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 11) | NC score | 0.409926 (rank : 10) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
WRIP1_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 12) | NC score | 0.375213 (rank : 12) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
RAD17_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 13) | NC score | 0.367985 (rank : 13) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6NXW6, O88934, O89024 | Gene names | Rad17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle checkpoint protein RAD17. | |||||
|
RAD17_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 14) | NC score | 0.334131 (rank : 14) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75943, O75714, Q7Z3S4, Q9UNK7, Q9UNR7, Q9UNR8, Q9UPF5 | Gene names | RAD17, R24L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle checkpoint protein RAD17 (hRad17) (RF-C/activator 1 homolog). | |||||
|
CDC6_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 15) | NC score | 0.146147 (rank : 15) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99741, Q8TB30 | Gene names | CDC6, CDC18L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division control protein 6 homolog (CDC6-related protein) (p62(cdc6)) (HsCDC6) (HsCDC18). | |||||
|
CLPX_HUMAN
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.100378 (rank : 17) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
CLPX_MOUSE
|
||||||
| θ value | 0.163984 (rank : 17) | NC score | 0.100500 (rank : 16) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JHS4, Q9WVD1 | Gene names | Clpx | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
NVL_HUMAN
|
||||||
| θ value | 0.163984 (rank : 18) | NC score | 0.075810 (rank : 20) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
NVL_MOUSE
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.074824 (rank : 21) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
YMEL1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.067347 (rank : 24) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96TA2, Q9H1Q0, Q9UMR9 | Gene names | YME1L1, FTSH1, YME1L | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin- associated metalloprotease) (PAMP). | |||||
|
YMEL1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.065971 (rank : 25) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
M10L1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.052950 (rank : 36) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BXT6, Q5TGD5, Q8NBD4, Q9NXW3, Q9UFB3, Q9UGX9 | Gene names | MOV10L1 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase Mov10l1 (EC 3.6.1.-) (Moloney leukemia virus 10-like protein 1) (MOV10-like 1). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.010565 (rank : 49) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MOD5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.048565 (rank : 42) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H3H1, Q6IAC9, Q96FJ3, Q96L45, Q9NXT7 | Gene names | TRIT1, IPT, MOD5 | |||
|
Domain Architecture |
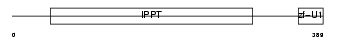
|
|||||
| Description | tRNA isopentenyltransferase, mitochondrial precursor (EC 2.5.1.8) (Isopentenyl-diphosphate:tRNA isopentenyltransferase) (IPP transferase) (IPTase) (IPPT) (hGRO1). | |||||
|
MOD5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.048884 (rank : 41) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80UN9, Q9D1H5 | Gene names | Trit1, Ipt | |||
|
Domain Architecture |
|
|||||
| Description | tRNA isopentenyltransferase, mitochondrial precursor (EC 2.5.1.8) (Isopentenyl-diphosphate:tRNA isopentenyltransferase) (IPP transferase) (IPTase) (IPPT). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.009640 (rank : 51) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
PRS10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.065663 (rank : 26) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P62333, P49719, Q6IBU3, Q92524 | Gene names | PSMC6, SUG2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS10_MOUSE
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.065663 (rank : 27) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P62334, P49719, Q3TKK1, Q810A6, Q92524, Q9CXH9 | Gene names | Psmc6, Sug2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
RUVB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.098365 (rank : 18) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y265, Q9BSX9 | Gene names | RUVBL1, NMP238, TIP49, TIP49A | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (Nuclear matrix protein 238) (NMP 238) (54 kDa erythrocyte cytosolic protein) (ECP-54) (TIP60-associated protein 54-alpha) (TAP54-alpha). | |||||
|
RUVB1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.098358 (rank : 19) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60122, O35753 | Gene names | Ruvbl1, Tip49, Tip49a | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (DNA helicase p50). | |||||
|
RUVB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.056238 (rank : 30) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y230, Q6PK27, Q9Y361 | Gene names | RUVBL2, TIP48, TIP49B | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (48 kDa TATA box-binding protein-interacting protein) (48 kDa TBP-interacting protein) (TIP49b) (Repressing pontin 52) (Reptin 52) (51 kDa erythrocyte cytosolic protein) (ECP-51) (TIP60-associated protein 54-beta) (TAP54-beta). | |||||
|
RUVB2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.056255 (rank : 29) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WTM5 | Gene names | Ruvbl2 | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (p47 protein). | |||||
|
ATPBB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.033032 (rank : 45) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VEJ1 | Gene names | Atpbd1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding domain 1 family member B. | |||||
|
CDC6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.072243 (rank : 22) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O89033, Q3TMD0, Q8C3S5, Q9CZT0 | Gene names | Cdc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division control protein 6 homolog (CDC6-related protein) (p62(cdc6)). | |||||
|
SMYD4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.023390 (rank : 47) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
ATD3B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.054818 (rank : 32) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.009818 (rank : 50) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
VPS4A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.044276 (rank : 43) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.044266 (rank : 44) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
MCM8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.009257 (rank : 52) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CWV1, Q80US2, Q80VI0 | Gene names | Mcm8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication licensing factor MCM8 (Minichromosome maintenance 8). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.007379 (rank : 53) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
PEX6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.055092 (rank : 31) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.003117 (rank : 55) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
RHG12_MOUSE
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.006786 (rank : 54) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
SETX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.015068 (rank : 48) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
THNSL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.028094 (rank : 46) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BH55 | Gene names | Thnsl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
ATAD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.050518 (rank : 40) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
BCS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.059926 (rank : 28) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y276, Q7Z2V7 | Gene names | BCS1L, BCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein) (H-BCS1). | |||||
|
BCS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.051720 (rank : 37) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CZP5 | Gene names | Bcs1l | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein). | |||||
|
LONM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.069659 (rank : 23) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
PEX6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.054542 (rank : 35) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PRS6A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.050557 (rank : 39) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17980, Q96HD3 | Gene names | PSMC3, TBP1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1) (Proteasome subunit P50). | |||||
|
PRS6A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.050582 (rank : 38) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
|
TERA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.054657 (rank : 34) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P55072, Q969G7 | Gene names | VCP | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
TERA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.054669 (rank : 33) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q01853 | Gene names | Vcp | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
RFC2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35250, P32846, Q9BU93 | Gene names | RFC2 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 2 (Replication factor C 40 kDa subunit) (RF-C 40 kDa subunit) (RFC40) (Activator 1 40 kDa subunit) (A1 40 kDa subunit). | |||||
|
RFC2_MOUSE
|
||||||
| NC score | 0.999332 (rank : 2) | θ value | 6.5317e-184 (rank : 2) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9WUK4 | Gene names | Rfc2 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 2 (Replication factor C 40 kDa subunit) (RF-C 40 kDa subunit) (RFC40) (Activator 1 40 kDa subunit) (A1 40 kDa subunit). | |||||
|
RFC5_HUMAN
|
||||||
| NC score | 0.928570 (rank : 3) | θ value | 1.0458e-56 (rank : 3) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P40937 | Gene names | RFC5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RFC5_MOUSE
|
||||||
| NC score | 0.928150 (rank : 4) | θ value | 1.97229e-55 (rank : 4) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D0F6 | Gene names | Rfc5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RFC4_HUMAN
|
||||||
| NC score | 0.912155 (rank : 5) | θ value | 8.57503e-51 (rank : 5) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P35249, Q6FHX7 | Gene names | RFC4 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 4 (Replication factor C 37 kDa subunit) (RF-C 37 kDa subunit) (RFC37) (Activator 1 37 kDa subunit) (A1 37 kDa subunit). | |||||
|
RFC4_MOUSE
|
||||||
| NC score | 0.909806 (rank : 6) | θ value | 3.04986e-48 (rank : 6) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99J62 | Gene names | Rfc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Replication factor C subunit 4. | |||||
|
RFC3_MOUSE
|
||||||
| NC score | 0.776148 (rank : 7) | θ value | 6.40375e-22 (rank : 8) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R323 | Gene names | Rfc3 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 3 (Replication factor C 38 kDa subunit) (RFC38) (Activator 1 38 kDa subunit) (A1 38 kDa subunit) (RF-C 38 kDa subunit). | |||||
|
RFC3_HUMAN
|
||||||
| NC score | 0.770951 (rank : 8) | θ value | 6.40375e-22 (rank : 7) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P40938, O15252 | Gene names | RFC3 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 3 (Replication factor C 38 kDa subunit) (RFC38) (Activator 1 38 kDa subunit) (A1 38 kDa subunit) (RF-C 38 kDa subunit). | |||||
|
RFC1_MOUSE
|
||||||
| NC score | 0.412339 (rank : 9) | θ value | 6.21693e-09 (rank : 9) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
WRIP1_MOUSE
|
||||||
| NC score | 0.409926 (rank : 10) | θ value | 2.36244e-08 (rank : 11) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
RFC1_HUMAN
|
||||||
| NC score | 0.398793 (rank : 11) | θ value | 8.11959e-09 (rank : 10) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
WRIP1_HUMAN
|
||||||
| NC score | 0.375213 (rank : 12) | θ value | 1.99992e-07 (rank : 12) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
RAD17_MOUSE
|
||||||
| NC score | 0.367985 (rank : 13) | θ value | 5.44631e-05 (rank : 13) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6NXW6, O88934, O89024 | Gene names | Rad17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle checkpoint protein RAD17. | |||||
|
RAD17_HUMAN
|
||||||
| NC score | 0.334131 (rank : 14) | θ value | 0.00134147 (rank : 14) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75943, O75714, Q7Z3S4, Q9UNK7, Q9UNR7, Q9UNR8, Q9UPF5 | Gene names | RAD17, R24L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle checkpoint protein RAD17 (hRad17) (RF-C/activator 1 homolog). | |||||
|
CDC6_HUMAN
|
||||||
| NC score | 0.146147 (rank : 15) | θ value | 0.0431538 (rank : 15) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99741, Q8TB30 | Gene names | CDC6, CDC18L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division control protein 6 homolog (CDC6-related protein) (p62(cdc6)) (HsCDC6) (HsCDC18). | |||||
|
CLPX_MOUSE
|
||||||
| NC score | 0.100500 (rank : 16) | θ value | 0.163984 (rank : 17) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JHS4, Q9WVD1 | Gene names | Clpx | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
CLPX_HUMAN
|
||||||
| NC score | 0.100378 (rank : 17) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
RUVB1_HUMAN
|
||||||
| NC score | 0.098365 (rank : 18) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y265, Q9BSX9 | Gene names | RUVBL1, NMP238, TIP49, TIP49A | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (Nuclear matrix protein 238) (NMP 238) (54 kDa erythrocyte cytosolic protein) (ECP-54) (TIP60-associated protein 54-alpha) (TAP54-alpha). | |||||
|
RUVB1_MOUSE
|
||||||
| NC score | 0.098358 (rank : 19) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60122, O35753 | Gene names | Ruvbl1, Tip49, Tip49a | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (DNA helicase p50). | |||||
|
NVL_HUMAN
|
||||||
| NC score | 0.075810 (rank : 20) | θ value | 0.163984 (rank : 18) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
NVL_MOUSE
|
||||||
| NC score | 0.074824 (rank : 21) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
CDC6_MOUSE
|
||||||
| NC score | 0.072243 (rank : 22) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O89033, Q3TMD0, Q8C3S5, Q9CZT0 | Gene names | Cdc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division control protein 6 homolog (CDC6-related protein) (p62(cdc6)). | |||||
|
LONM_HUMAN
|
||||||
| NC score | 0.069659 (rank : 23) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
YMEL1_HUMAN
|
||||||
| NC score | 0.067347 (rank : 24) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96TA2, Q9H1Q0, Q9UMR9 | Gene names | YME1L1, FTSH1, YME1L | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin- associated metalloprotease) (PAMP). | |||||
|
YMEL1_MOUSE
|
||||||
| NC score | 0.065971 (rank : 25) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
PRS10_HUMAN
|
||||||
| NC score | 0.065663 (rank : 26) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P62333, P49719, Q6IBU3, Q92524 | Gene names | PSMC6, SUG2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS10_MOUSE
|
||||||
| NC score | 0.065663 (rank : 27) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P62334, P49719, Q3TKK1, Q810A6, Q92524, Q9CXH9 | Gene names | Psmc6, Sug2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
BCS1_HUMAN
|
||||||
| NC score | 0.059926 (rank : 28) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y276, Q7Z2V7 | Gene names | BCS1L, BCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein) (H-BCS1). | |||||
|
RUVB2_MOUSE
|
||||||
| NC score | 0.056255 (rank : 29) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WTM5 | Gene names | Ruvbl2 | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (p47 protein). | |||||
|
RUVB2_HUMAN
|
||||||
| NC score | 0.056238 (rank : 30) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y230, Q6PK27, Q9Y361 | Gene names | RUVBL2, TIP48, TIP49B | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (48 kDa TATA box-binding protein-interacting protein) (48 kDa TBP-interacting protein) (TIP49b) (Repressing pontin 52) (Reptin 52) (51 kDa erythrocyte cytosolic protein) (ECP-51) (TIP60-associated protein 54-beta) (TAP54-beta). | |||||
|
PEX6_MOUSE
|
||||||
| NC score | 0.055092 (rank : 31) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
ATD3B_HUMAN
|
||||||
| NC score | 0.054818 (rank : 32) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
TERA_MOUSE
|
||||||
| NC score | 0.054669 (rank : 33) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q01853 | Gene names | Vcp | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
TERA_HUMAN
|
||||||
| NC score | 0.054657 (rank : 34) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P55072, Q969G7 | Gene names | VCP | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
PEX6_HUMAN
|
||||||
| NC score | 0.054542 (rank : 35) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
M10L1_HUMAN
|
||||||
| NC score | 0.052950 (rank : 36) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BXT6, Q5TGD5, Q8NBD4, Q9NXW3, Q9UFB3, Q9UGX9 | Gene names | MOV10L1 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase Mov10l1 (EC 3.6.1.-) (Moloney leukemia virus 10-like protein 1) (MOV10-like 1). | |||||
|
BCS1_MOUSE
|
||||||
| NC score | 0.051720 (rank : 37) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CZP5 | Gene names | Bcs1l | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein). | |||||
|
PRS6A_MOUSE
|
||||||
| NC score | 0.050582 (rank : 38) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
|
PRS6A_HUMAN
|
||||||
| NC score | 0.050557 (rank : 39) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17980, Q96HD3 | Gene names | PSMC3, TBP1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1) (Proteasome subunit P50). | |||||
|
ATAD3_MOUSE
|
||||||
| NC score | 0.050518 (rank : 40) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
MOD5_MOUSE
|
||||||
| NC score | 0.048884 (rank : 41) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80UN9, Q9D1H5 | Gene names | Trit1, Ipt | |||
|
Domain Architecture |

|
|||||
| Description | tRNA isopentenyltransferase, mitochondrial precursor (EC 2.5.1.8) (Isopentenyl-diphosphate:tRNA isopentenyltransferase) (IPP transferase) (IPTase) (IPPT). | |||||
|
MOD5_HUMAN
|
||||||
| NC score | 0.048565 (rank : 42) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H3H1, Q6IAC9, Q96FJ3, Q96L45, Q9NXT7 | Gene names | TRIT1, IPT, MOD5 | |||
|
Domain Architecture |

|
|||||
| Description | tRNA isopentenyltransferase, mitochondrial precursor (EC 2.5.1.8) (Isopentenyl-diphosphate:tRNA isopentenyltransferase) (IPP transferase) (IPTase) (IPPT) (hGRO1). | |||||
|
VPS4A_HUMAN
|
||||||
| NC score | 0.044276 (rank : 43) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| NC score | 0.044266 (rank : 44) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
ATPBB_MOUSE
|
||||||
| NC score | 0.033032 (rank : 45) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VEJ1 | Gene names | Atpbd1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding domain 1 family member B. | |||||
|
THNSL_MOUSE
|
||||||
| NC score | 0.028094 (rank : 46) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BH55 | Gene names | Thnsl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
SMYD4_HUMAN
|
||||||
| NC score | 0.023390 (rank : 47) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
SETX_HUMAN
|
||||||
| NC score | 0.015068 (rank : 48) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.010565 (rank : 49) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.009818 (rank : 50) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.009640 (rank : 51) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MCM8_MOUSE
|
||||||
| NC score | 0.009257 (rank : 52) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CWV1, Q80US2, Q80VI0 | Gene names | Mcm8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication licensing factor MCM8 (Minichromosome maintenance 8). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.007379 (rank : 53) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
RHG12_MOUSE
|
||||||
| NC score | 0.006786 (rank : 54) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.003117 (rank : 55) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||