Please be patient as the page loads
|
NAB1_MOUSE
|
||||||
| SwissProt Accessions | Q61122 | Gene names | Nab1 | |||
|
Domain Architecture |
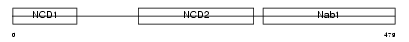
|
|||||
| Description | NGFI-A-binding protein 1 (EGR-1-binding protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NAB1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.991677 (rank : 2) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13506, O75383, O75384, Q6GTU1, Q9UEV1 | Gene names | NAB1 | |||
|
Domain Architecture |
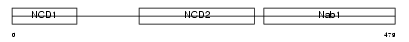
|
|||||
| Description | NGFI-A-binding protein 1 (EGR-1-binding protein 1) (Transcriptional regulatory protein p54). | |||||
|
NAB1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61122 | Gene names | Nab1 | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 1 (EGR-1-binding protein 1). | |||||
|
NAB2_HUMAN
|
||||||
| θ value | 8.49582e-83 (rank : 3) | NC score | 0.921775 (rank : 3) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15742, O76006, Q14797 | Gene names | NAB2, MADER | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 2 (EGR-1-binding protein 2) (Melanoma- associated delayed early response protein) (Protein MADER). | |||||
|
NAB2_MOUSE
|
||||||
| θ value | 2.02684e-76 (rank : 4) | NC score | 0.917979 (rank : 4) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61127 | Gene names | Nab2 | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 2 (EGR-1-binding protein 2). | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 5) | NC score | 0.015129 (rank : 10) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 6) | NC score | 0.015204 (rank : 9) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
CHD1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 7) | NC score | 0.014697 (rank : 11) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 8) | NC score | 0.022351 (rank : 5) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
K2C7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 9) | NC score | 0.006755 (rank : 16) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08729, Q92676, Q9BUD8, Q9Y3R7 | Gene names | KRT7, SCL | |||
|
Domain Architecture |
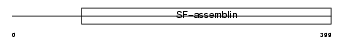
|
|||||
| Description | Keratin, type II cytoskeletal 7 (Cytokeratin-7) (CK-7) (Keratin-7) (K7) (Sarcolectin). | |||||
|
MERL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 10) | NC score | 0.013100 (rank : 13) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35240, O95683, Q8WUJ2, Q969N0, Q969Q3, Q96T30, Q96T31, Q96T32, Q96T33, Q9BTW3, Q9UNG9, Q9UNH3, Q9UNH4 | Gene names | NF2, SCH | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin) (Schwannomerlin). | |||||
|
MERL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 11) | NC score | 0.013138 (rank : 12) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P46662, Q8BR03 | Gene names | Nf2, Nf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin). | |||||
|
PRS6A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 12) | NC score | 0.018675 (rank : 7) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17980, Q96HD3 | Gene names | PSMC3, TBP1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1) (Proteasome subunit P50). | |||||
|
PRS6A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.018726 (rank : 6) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
|
ABCA7_MOUSE
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.016347 (rank : 8) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91V24, Q9JL36 | Gene names | Abca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 7. | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.011923 (rank : 14) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
PCDGF_HUMAN
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.003286 (rank : 19) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5G1, Q9Y5C7 | Gene names | PCDHGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B3 precursor (PCDH-gamma-B3). | |||||
|
SPTN4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.006348 (rank : 17) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
MEFV_MOUSE
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.010064 (rank : 15) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJ26 | Gene names | Mefv | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 19) | NC score | 0.003647 (rank : 18) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
NAB1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61122 | Gene names | Nab1 | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 1 (EGR-1-binding protein 1). | |||||
|
NAB1_HUMAN
|
||||||
| NC score | 0.991677 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13506, O75383, O75384, Q6GTU1, Q9UEV1 | Gene names | NAB1 | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 1 (EGR-1-binding protein 1) (Transcriptional regulatory protein p54). | |||||
|
NAB2_HUMAN
|
||||||
| NC score | 0.921775 (rank : 3) | θ value | 8.49582e-83 (rank : 3) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15742, O76006, Q14797 | Gene names | NAB2, MADER | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 2 (EGR-1-binding protein 2) (Melanoma- associated delayed early response protein) (Protein MADER). | |||||
|
NAB2_MOUSE
|
||||||
| NC score | 0.917979 (rank : 4) | θ value | 2.02684e-76 (rank : 4) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61127 | Gene names | Nab2 | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 2 (EGR-1-binding protein 2). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.022351 (rank : 5) | θ value | 3.0926 (rank : 8) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
PRS6A_MOUSE
|
||||||
| NC score | 0.018726 (rank : 6) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
|
PRS6A_HUMAN
|
||||||
| NC score | 0.018675 (rank : 7) | θ value | 3.0926 (rank : 12) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17980, Q96HD3 | Gene names | PSMC3, TBP1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1) (Proteasome subunit P50). | |||||
|
ABCA7_MOUSE
|
||||||
| NC score | 0.016347 (rank : 8) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91V24, Q9JL36 | Gene names | Abca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 7. | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.015204 (rank : 9) | θ value | 2.36792 (rank : 6) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.015129 (rank : 10) | θ value | 2.36792 (rank : 5) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD1_MOUSE
|
||||||
| NC score | 0.014697 (rank : 11) | θ value | 3.0926 (rank : 7) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
MERL_MOUSE
|
||||||
| NC score | 0.013138 (rank : 12) | θ value | 3.0926 (rank : 11) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P46662, Q8BR03 | Gene names | Nf2, Nf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin). | |||||
|
MERL_HUMAN
|
||||||
| NC score | 0.013100 (rank : 13) | θ value | 3.0926 (rank : 10) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35240, O95683, Q8WUJ2, Q969N0, Q969Q3, Q96T30, Q96T31, Q96T32, Q96T33, Q9BTW3, Q9UNG9, Q9UNH3, Q9UNH4 | Gene names | NF2, SCH | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin) (Schwannomerlin). | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.011923 (rank : 14) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
MEFV_MOUSE
|
||||||
| NC score | 0.010064 (rank : 15) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJ26 | Gene names | Mefv | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
K2C7_HUMAN
|
||||||
| NC score | 0.006755 (rank : 16) | θ value | 3.0926 (rank : 9) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08729, Q92676, Q9BUD8, Q9Y3R7 | Gene names | KRT7, SCL | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 7 (Cytokeratin-7) (CK-7) (Keratin-7) (K7) (Sarcolectin). | |||||
|
SPTN4_HUMAN
|
||||||
| NC score | 0.006348 (rank : 17) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.003647 (rank : 18) | θ value | 8.99809 (rank : 19) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PCDGF_HUMAN
|
||||||
| NC score | 0.003286 (rank : 19) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5G1, Q9Y5C7 | Gene names | PCDHGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B3 precursor (PCDH-gamma-B3). | |||||