Please be patient as the page loads
|
KTNA1_MOUSE
|
||||||
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
KTNA1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.997142 (rank : 2) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | O75449, Q5TFA8, Q5TFA9, Q86VN2, Q9NU52 | Gene names | KATNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin). | |||||
|
KTNA1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 123 | |
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
|
KATL1_HUMAN
|
||||||
| θ value | 1.19313e-177 (rank : 3) | NC score | 0.993928 (rank : 4) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9BW62 | Gene names | KATNAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
KATL1_MOUSE
|
||||||
| θ value | 1.31915e-176 (rank : 4) | NC score | 0.994086 (rank : 3) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8K0T4 | Gene names | Katnal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
VPS4A_HUMAN
|
||||||
| θ value | 2.56992e-63 (rank : 5) | NC score | 0.956185 (rank : 11) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| θ value | 2.56992e-63 (rank : 6) | NC score | 0.956155 (rank : 12) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
SPAST_HUMAN
|
||||||
| θ value | 6.32992e-62 (rank : 7) | NC score | 0.961189 (rank : 5) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UBP0, Q9UPR9 | Gene names | SPAST, KIAA1083, SPG4 | |||
|
Domain Architecture |

|
|||||
| Description | Spastin. | |||||
|
SPAST_MOUSE
|
||||||
| θ value | 6.32992e-62 (rank : 8) | NC score | 0.959766 (rank : 6) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
VPS4B_MOUSE
|
||||||
| θ value | 1.41016e-61 (rank : 9) | NC score | 0.957988 (rank : 10) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4B_HUMAN
|
||||||
| θ value | 4.10295e-61 (rank : 10) | NC score | 0.958455 (rank : 9) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
ATAD1_HUMAN
|
||||||
| θ value | 9.81109e-47 (rank : 11) | NC score | 0.959189 (rank : 7) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8NBU5, Q6P4B9, Q8N3G1, Q8WYR9, Q969Y3 | Gene names | ATAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
ATAD1_MOUSE
|
||||||
| θ value | 9.81109e-47 (rank : 12) | NC score | 0.959189 (rank : 8) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9D5T0, Q3U8V2, Q9D7A4 | Gene names | Atad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
FIGN_MOUSE
|
||||||
| θ value | 3.72821e-46 (rank : 13) | NC score | 0.926348 (rank : 17) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
FIGN_HUMAN
|
||||||
| θ value | 1.85029e-45 (rank : 14) | NC score | 0.921935 (rank : 22) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
TERA_HUMAN
|
||||||
| θ value | 6.58091e-43 (rank : 15) | NC score | 0.927489 (rank : 13) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P55072, Q969G7 | Gene names | VCP | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
TERA_MOUSE
|
||||||
| θ value | 6.58091e-43 (rank : 16) | NC score | 0.927447 (rank : 14) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q01853 | Gene names | Vcp | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
PRS8_HUMAN
|
||||||
| θ value | 2.76489e-41 (rank : 17) | NC score | 0.925077 (rank : 18) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P62195, O35051, O43208, P47210, P52915, P52916 | Gene names | PSMC5, SUG1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (Thyroid hormone receptor- interacting protein 1) (TRIP1). | |||||
|
PRS8_MOUSE
|
||||||
| θ value | 2.76489e-41 (rank : 18) | NC score | 0.925077 (rank : 19) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P62196, O35051, P47210, P52915, P52916, Q3UL51, Q9CWN5 | Gene names | Psmc5, Sug1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (mSUG1). | |||||
|
PRS10_HUMAN
|
||||||
| θ value | 1.51715e-39 (rank : 19) | NC score | 0.926819 (rank : 15) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P62333, P49719, Q6IBU3, Q92524 | Gene names | PSMC6, SUG2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS10_MOUSE
|
||||||
| θ value | 1.51715e-39 (rank : 20) | NC score | 0.926819 (rank : 16) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P62334, P49719, Q3TKK1, Q810A6, Q92524, Q9CXH9 | Gene names | Psmc6, Sug2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS4_HUMAN
|
||||||
| θ value | 1.98146e-39 (rank : 21) | NC score | 0.921292 (rank : 23) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P62191, P49014, Q03527, Q6IAW0, Q96AZ3 | Gene names | PSMC1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS4_MOUSE
|
||||||
| θ value | 1.98146e-39 (rank : 22) | NC score | 0.921292 (rank : 24) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P62192, P49014, Q03527, Q96AZ3 | Gene names | Psmc1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
NVL_MOUSE
|
||||||
| θ value | 3.37985e-39 (rank : 23) | NC score | 0.917785 (rank : 26) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
NVL_HUMAN
|
||||||
| θ value | 7.52953e-39 (rank : 24) | NC score | 0.917151 (rank : 27) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PRS6A_HUMAN
|
||||||
| θ value | 8.32485e-38 (rank : 25) | NC score | 0.922671 (rank : 21) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P17980, Q96HD3 | Gene names | PSMC3, TBP1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1) (Proteasome subunit P50). | |||||
|
PRS6A_MOUSE
|
||||||
| θ value | 8.32485e-38 (rank : 26) | NC score | 0.922854 (rank : 20) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
|
PEX6_MOUSE
|
||||||
| θ value | 5.39604e-37 (rank : 27) | NC score | 0.913375 (rank : 33) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PEX6_HUMAN
|
||||||
| θ value | 2.67802e-36 (rank : 28) | NC score | 0.913518 (rank : 32) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PEX1_HUMAN
|
||||||
| θ value | 2.12192e-33 (rank : 29) | NC score | 0.919690 (rank : 25) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
YMEL1_MOUSE
|
||||||
| θ value | 2.12192e-33 (rank : 30) | NC score | 0.901291 (rank : 34) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
AFG31_MOUSE
|
||||||
| θ value | 1.37539e-32 (rank : 31) | NC score | 0.896499 (rank : 36) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q920A7 | Gene names | Afg3l1 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 1 (EC 3.4.24.-). | |||||
|
PRS7_HUMAN
|
||||||
| θ value | 4.00176e-32 (rank : 32) | NC score | 0.916733 (rank : 28) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P35998 | Gene names | PSMC2, MSS1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS7_MOUSE
|
||||||
| θ value | 4.00176e-32 (rank : 33) | NC score | 0.916724 (rank : 29) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P46471, O08531, Q3TGX5, Q9DBA1 | Gene names | Psmc2, Mss1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
ATAD2_HUMAN
|
||||||
| θ value | 8.91499e-32 (rank : 34) | NC score | 0.851530 (rank : 43) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
PRS6B_HUMAN
|
||||||
| θ value | 1.16434e-31 (rank : 35) | NC score | 0.915946 (rank : 30) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P43686, Q96FV5, Q9UBM3, Q9UEX3 | Gene names | PSMC4, TBP7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7). | |||||
|
ATAD2_MOUSE
|
||||||
| θ value | 1.98606e-31 (rank : 36) | NC score | 0.868947 (rank : 42) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
PRS6B_MOUSE
|
||||||
| θ value | 5.77852e-31 (rank : 37) | NC score | 0.914773 (rank : 31) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P54775 | Gene names | Psmc4, Tbp7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7) (CIP21). | |||||
|
AFG32_HUMAN
|
||||||
| θ value | 2.19584e-30 (rank : 38) | NC score | 0.890656 (rank : 38) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9Y4W6 | Gene names | AFG3L2 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-) (Paraplegin-like protein). | |||||
|
AFG32_MOUSE
|
||||||
| θ value | 2.86786e-30 (rank : 39) | NC score | 0.890492 (rank : 39) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8JZQ2 | Gene names | Afg3l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-). | |||||
|
YMEL1_HUMAN
|
||||||
| θ value | 1.4233e-29 (rank : 40) | NC score | 0.897769 (rank : 35) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96TA2, Q9H1Q0, Q9UMR9 | Gene names | YME1L1, FTSH1, YME1L | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin- associated metalloprotease) (PAMP). | |||||
|
SPG7_HUMAN
|
||||||
| θ value | 3.17079e-29 (rank : 41) | NC score | 0.896205 (rank : 37) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UQ90, O75756, Q96IB0 | Gene names | SPG7, CAR, CMAR, PGN | |||
|
Domain Architecture |

|
|||||
| Description | Paraplegin (EC 3.4.24.-) (Spastic paraplegia protein 7). | |||||
|
NSF_HUMAN
|
||||||
| θ value | 3.75424e-22 (rank : 42) | NC score | 0.876233 (rank : 40) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P46459, Q9UKZ2 | Gene names | NSF | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein). | |||||
|
NSF_MOUSE
|
||||||
| θ value | 3.75424e-22 (rank : 43) | NC score | 0.875738 (rank : 41) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P46460 | Gene names | Nsf, Skd2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein) (SKD2 protein). | |||||
|
ATD3B_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 44) | NC score | 0.622660 (rank : 47) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
ATAD3_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 45) | NC score | 0.614020 (rank : 48) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
ATD3A_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 46) | NC score | 0.638257 (rank : 46) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9NVI7, Q8N275, Q96A50 | Gene names | ATAD3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3A. | |||||
|
TRP13_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 47) | NC score | 0.713959 (rank : 44) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q3UA06, Q3UQG6, Q9CWW8 | Gene names | Trip13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13). | |||||
|
TRP13_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 48) | NC score | 0.712445 (rank : 45) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q15645, O15324 | Gene names | TRIP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13) (Human papillomavirus type 16 E1 protein- binding protein) (HPV16 E1 protein-binding protein) (16E1-BP). | |||||
|
CLPX_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 49) | NC score | 0.425665 (rank : 51) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
CLPX_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 50) | NC score | 0.422852 (rank : 52) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JHS4, Q9WVD1 | Gene names | Clpx | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
LIMA1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 51) | NC score | 0.025879 (rank : 75) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ERG0, Q9ERG1 | Gene names | Lima1, D15Ertd366e, Eplin | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm) (mEPLIN). | |||||
|
FOXM1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 52) | NC score | 0.020471 (rank : 78) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q08050, O43258, O43259, O43260, Q4ZGG7, Q9BRL2 | Gene names | FOXM1, FKHL16, HFH11, MPP2, WIN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead-related protein FKHL16) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/fork-head homolog 11) (HFH-11) (Winged helix factor from INS-1 cells) (M-phase phosphoprotein 2) (MPM-2 reactive phosphoprotein 2) (Transcription factor Trident). | |||||
|
RUVB1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 53) | NC score | 0.324473 (rank : 54) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y265, Q9BSX9 | Gene names | RUVBL1, NMP238, TIP49, TIP49A | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (Nuclear matrix protein 238) (NMP 238) (54 kDa erythrocyte cytosolic protein) (ECP-54) (TIP60-associated protein 54-alpha) (TAP54-alpha). | |||||
|
RUVB1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 54) | NC score | 0.324434 (rank : 55) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P60122, O35753 | Gene names | Ruvbl1, Tip49, Tip49a | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (DNA helicase p50). | |||||
|
BANK1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 55) | NC score | 0.038822 (rank : 71) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80VH0, Q3U178, Q8BRV6, Q8BRY8 | Gene names | Bank1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell scaffold protein with ankyrin repeats (Protein AVIEF). | |||||
|
LONM_HUMAN
|
||||||
| θ value | 0.125558 (rank : 56) | NC score | 0.413642 (rank : 53) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
CIR_MOUSE
|
||||||
| θ value | 0.21417 (rank : 57) | NC score | 0.031967 (rank : 73) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
SYYM_MOUSE
|
||||||
| θ value | 0.21417 (rank : 58) | NC score | 0.030369 (rank : 74) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BYL4, Q6PAH7 | Gene names | Yars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.1) (Tyrosine--tRNA ligase) (TyrRS). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 59) | NC score | 0.008704 (rank : 96) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 0.365318 (rank : 60) | NC score | -0.004121 (rank : 123) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
BANK1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 61) | NC score | 0.032564 (rank : 72) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NDB2, Q8N5K8, Q8NB56, Q8WYN5, Q9NWP2 | Gene names | BANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell scaffold protein with ankyrin repeats. | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 0.47712 (rank : 62) | NC score | 0.009474 (rank : 93) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
WRIP1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 63) | NC score | 0.230738 (rank : 56) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
SFR12_HUMAN
|
||||||
| θ value | 0.62314 (rank : 64) | NC score | 0.013185 (rank : 86) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 65) | NC score | 0.014807 (rank : 82) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
THNSL_MOUSE
|
||||||
| θ value | 0.62314 (rank : 66) | NC score | 0.131743 (rank : 58) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BH55 | Gene names | Thnsl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
WRIP1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 67) | NC score | 0.229241 (rank : 57) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
CTCF_HUMAN
|
||||||
| θ value | 0.813845 (rank : 68) | NC score | -0.004244 (rank : 124) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49711 | Gene names | CTCF | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor CTCF (CCCTC-binding factor) (CTCFL paralog) (11-zinc finger protein). | |||||
|
RFC4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 69) | NC score | 0.108797 (rank : 64) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35249, Q6FHX7 | Gene names | RFC4 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 4 (Replication factor C 37 kDa subunit) (RF-C 37 kDa subunit) (RFC37) (Activator 1 37 kDa subunit) (A1 37 kDa subunit). | |||||
|
SAHH3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 70) | NC score | 0.012748 (rank : 87) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96HN2, O94917 | Gene names | KIAA0828 | |||
|
Domain Architecture |

|
|||||
| Description | Putative adenosylhomocysteinase 3 (EC 3.3.1.1) (S-adenosyl-L- homocysteine hydrolase 3) (AdoHcyase 3). | |||||
|
IMUP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 71) | NC score | 0.071806 (rank : 68) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9GZP8, Q96H58, Q9HCR4 | Gene names | IMUP, C19orf33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immortalization-up-regulated protein (Hepatocyte growth factor activator inhibitor type 2-related small protein) (HAI-2-related small protein) (H2RSP). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 72) | NC score | 0.011684 (rank : 89) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TM131_MOUSE
|
||||||
| θ value | 1.06291 (rank : 73) | NC score | 0.018468 (rank : 79) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O70472 | Gene names | Tmem131, D1Bwg0491e, Rw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 131 (Protein RW1). | |||||
|
PO6F2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.002327 (rank : 115) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
RFC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.120103 (rank : 62) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
RFC1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.131007 (rank : 59) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
MAGI1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.003376 (rank : 112) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
THOC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.020650 (rank : 77) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
APC_MOUSE
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.009745 (rank : 91) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.016057 (rank : 81) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
RFC4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.097165 (rank : 66) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99J62 | Gene names | Rfc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Replication factor C subunit 4. | |||||
|
THNSL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.119957 (rank : 63) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IYQ7, Q5VV21 | Gene names | THNSL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
AP3D1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.005742 (rank : 104) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
M4K4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | -0.005119 (rank : 125) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
SMBP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.018451 (rank : 80) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P40694 | Gene names | Ighmbp2, Smbp-2, Smbp2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Cardiac transcription factor 1) (CATF1). | |||||
|
STK6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | -0.005186 (rank : 126) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14965, O60445, O75873, Q9BQD6, Q9UPG5 | Gene names | STK6, AIK, ARK1, AURA, BTAK, STK15 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 6 (EC 2.7.11.1) (Serine/threonine kinase 15) (Aurora/IPL1-related kinase 1) (Aurora-related kinase 1) (hARK1) (Aurora-A) (Breast-tumor-amplified kinase). | |||||
|
SYYM_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.023580 (rank : 76) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2Z4, Q9H817 | Gene names | YARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.1) (Tyrosine--tRNA ligase) (TyrRS). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.006995 (rank : 100) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
BCS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.570372 (rank : 49) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y276, Q7Z2V7 | Gene names | BCS1L, BCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein) (H-BCS1). | |||||
|
RTKN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.014788 (rank : 83) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C6B2, Q61192, Q8VIG7 | Gene names | Rtkn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rhotekin. | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.011612 (rank : 90) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
TGM5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.003603 (rank : 111) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D7I9, Q3V1F9 | Gene names | Tgm5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-glutamine gamma-glutamyltransferase 5 (EC 2.3.2.13) (Transglutaminase-5) (TGase 5). | |||||
|
TSH1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.006147 (rank : 103) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
UTRO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | -0.000556 (rank : 120) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
BARX2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | -0.000016 (rank : 118) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08686 | Gene names | Barx2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein BarH-like 2. | |||||
|
CPXM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.002165 (rank : 116) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N436 | Gene names | CPXM2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
|
DYH5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.009203 (rank : 94) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
FA44A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.007605 (rank : 98) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.004005 (rank : 109) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
SF01_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.007575 (rank : 99) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.014711 (rank : 84) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
BCS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.559813 (rank : 50) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9CZP5 | Gene names | Bcs1l | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.014262 (rank : 85) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
CBLB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.004919 (rank : 107) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3TTA7 | Gene names | Cblb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase CBL-B (EC 6.3.2.-) (Signal transduction protein CBL-B) (SH3-binding protein CBL-B) (Casitas B-lineage lymphoma proto-oncogene b). | |||||
|
GIT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.000293 (rank : 117) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y2X7, Q86SS0, Q9BRJ4 | Gene names | GIT1 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase- interactor 1) (GRK-interacting protein 1) (Cool-associated and tyrosine-phosphorylated protein 1) (Cat-1). | |||||
|
K0427_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.006312 (rank : 101) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43310, Q8IVD5 | Gene names | KIAA0427 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0427. | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | -0.003399 (rank : 121) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
PRP4B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | -0.004073 (rank : 122) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1030 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13523, Q8TDP2, Q96QT7, Q9UEE6 | Gene names | PRPF4B, KIAA0536, PRP4, PRP4H, PRP4K | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (PRP4 kinase). | |||||
|
RU17_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.002883 (rank : 114) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08621, P78493, P78494, Q15364, Q15686, Q15687, Q15689, Q99377, Q9UE45, Q9UE46, Q9UE47, Q9UE48, Q9UFQ6 | Gene names | SNRP70, RPU1, U1AP1 | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 snRNP 70 kDa) (snRNP70) (U1-70K). | |||||
|
RU17_MOUSE
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.003017 (rank : 113) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62376, Q3UIW4 | Gene names | Snrp70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 SNRNP 70 kDa) (snRNP70). | |||||
|
SF01_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.007650 (rank : 97) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64213, O08817, P70167, Q61454, Q921Z4 | Gene names | Sf1, Zfm1, Zfp162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (mZFM) (Zinc finger gene in MEN1 locus) (Mammalian branch point- binding protein mBBP) (BBP) (CW17). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.012062 (rank : 88) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TBCD5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.005420 (rank : 106) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92609 | Gene names | TBC1D5, KIAA0210 | |||
|
Domain Architecture |
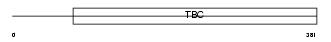
|
|||||
| Description | TBC1 domain family member 5. | |||||
|
TNNT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.004604 (rank : 108) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P45379, O60214, Q99596, Q99597, Q9UM96 | Gene names | TNNT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
TNNT2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.003681 (rank : 110) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P50752, Q64360, Q64377 | Gene names | Tnnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
DEK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.005488 (rank : 105) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
DMD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | -0.000489 (rank : 119) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
KAD6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.107701 (rank : 65) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
PP14D_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.006242 (rank : 102) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NXH3 | Gene names | PPP1R14D, GBPI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 14D (Gastrointestinal and brain-specific PP1-inhibitory protein 1) (GBPI-1). | |||||
|
RUVB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.123910 (rank : 60) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y230, Q6PK27, Q9Y361 | Gene names | RUVBL2, TIP48, TIP49B | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (48 kDa TATA box-binding protein-interacting protein) (48 kDa TBP-interacting protein) (TIP49b) (Repressing pontin 52) (Reptin 52) (51 kDa erythrocyte cytosolic protein) (ECP-51) (TIP60-associated protein 54-beta) (TAP54-beta). | |||||
|
RUVB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.120386 (rank : 61) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WTM5 | Gene names | Ruvbl2 | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (p47 protein). | |||||
|
SAHH3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.008963 (rank : 95) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q68FL4, Q8BIH1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative adenosylhomocysteinase 3 (EC 3.3.1.1) (S-adenosyl-L- homocysteine hydrolase 3) (AdoHcyase 3). | |||||
|
SPTA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.009503 (rank : 92) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NHX4, Q86WX5, Q8N9Y6 | Gene names | SPATA3, TSARG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 3 (Testis spermatocyte apoptosis- related protein 1) (Testis and spermatogenesis cell-related protein 1). | |||||
|
KAD6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.082420 (rank : 67) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VCP8 | Gene names | Ak6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
RFC5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.060017 (rank : 69) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P40937 | Gene names | RFC5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RFC5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.059264 (rank : 70) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9D0F6 | Gene names | Rfc5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
KTNA1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 123 | |
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
|
KTNA1_HUMAN
|
||||||
| NC score | 0.997142 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | O75449, Q5TFA8, Q5TFA9, Q86VN2, Q9NU52 | Gene names | KATNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin). | |||||
|
KATL1_MOUSE
|
||||||
| NC score | 0.994086 (rank : 3) | θ value | 1.31915e-176 (rank : 4) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8K0T4 | Gene names | Katnal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
KATL1_HUMAN
|
||||||
| NC score | 0.993928 (rank : 4) | θ value | 1.19313e-177 (rank : 3) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9BW62 | Gene names | KATNAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
SPAST_HUMAN
|
||||||
| NC score | 0.961189 (rank : 5) | θ value | 6.32992e-62 (rank : 7) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UBP0, Q9UPR9 | Gene names | SPAST, KIAA1083, SPG4 | |||
|
Domain Architecture |

|
|||||
| Description | Spastin. | |||||
|
SPAST_MOUSE
|
||||||
| NC score | 0.959766 (rank : 6) | θ value | 6.32992e-62 (rank : 8) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
ATAD1_HUMAN
|
||||||
| NC score | 0.959189 (rank : 7) | θ value | 9.81109e-47 (rank : 11) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8NBU5, Q6P4B9, Q8N3G1, Q8WYR9, Q969Y3 | Gene names | ATAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
ATAD1_MOUSE
|
||||||
| NC score | 0.959189 (rank : 8) | θ value | 9.81109e-47 (rank : 12) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9D5T0, Q3U8V2, Q9D7A4 | Gene names | Atad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
VPS4B_HUMAN
|
||||||
| NC score | 0.958455 (rank : 9) | θ value | 4.10295e-61 (rank : 10) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4B_MOUSE
|
||||||
| NC score | 0.957988 (rank : 10) | θ value | 1.41016e-61 (rank : 9) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4A_HUMAN
|
||||||
| NC score | 0.956185 (rank : 11) | θ value | 2.56992e-63 (rank : 5) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| NC score | 0.956155 (rank : 12) | θ value | 2.56992e-63 (rank : 6) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
TERA_HUMAN
|
||||||
| NC score | 0.927489 (rank : 13) | θ value | 6.58091e-43 (rank : 15) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P55072, Q969G7 | Gene names | VCP | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
TERA_MOUSE
|
||||||
| NC score | 0.927447 (rank : 14) | θ value | 6.58091e-43 (rank : 16) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q01853 | Gene names | Vcp | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
PRS10_HUMAN
|
||||||
| NC score | 0.926819 (rank : 15) | θ value | 1.51715e-39 (rank : 19) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P62333, P49719, Q6IBU3, Q92524 | Gene names | PSMC6, SUG2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS10_MOUSE
|
||||||
| NC score | 0.926819 (rank : 16) | θ value | 1.51715e-39 (rank : 20) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P62334, P49719, Q3TKK1, Q810A6, Q92524, Q9CXH9 | Gene names | Psmc6, Sug2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
FIGN_MOUSE
|
||||||
| NC score | 0.926348 (rank : 17) | θ value | 3.72821e-46 (rank : 13) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
PRS8_HUMAN
|
||||||
| NC score | 0.925077 (rank : 18) | θ value | 2.76489e-41 (rank : 17) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P62195, O35051, O43208, P47210, P52915, P52916 | Gene names | PSMC5, SUG1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (Thyroid hormone receptor- interacting protein 1) (TRIP1). | |||||
|
PRS8_MOUSE
|
||||||
| NC score | 0.925077 (rank : 19) | θ value | 2.76489e-41 (rank : 18) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P62196, O35051, P47210, P52915, P52916, Q3UL51, Q9CWN5 | Gene names | Psmc5, Sug1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (mSUG1). | |||||
|
PRS6A_MOUSE
|
||||||
| NC score | 0.922854 (rank : 20) | θ value | 8.32485e-38 (rank : 26) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
|
PRS6A_HUMAN
|
||||||
| NC score | 0.922671 (rank : 21) | θ value | 8.32485e-38 (rank : 25) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P17980, Q96HD3 | Gene names | PSMC3, TBP1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1) (Proteasome subunit P50). | |||||
|
FIGN_HUMAN
|
||||||
| NC score | 0.921935 (rank : 22) | θ value | 1.85029e-45 (rank : 14) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
PRS4_HUMAN
|
||||||
| NC score | 0.921292 (rank : 23) | θ value | 1.98146e-39 (rank : 21) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P62191, P49014, Q03527, Q6IAW0, Q96AZ3 | Gene names | PSMC1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS4_MOUSE
|
||||||
| NC score | 0.921292 (rank : 24) | θ value | 1.98146e-39 (rank : 22) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P62192, P49014, Q03527, Q96AZ3 | Gene names | Psmc1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PEX1_HUMAN
|
||||||
| NC score | 0.919690 (rank : 25) | θ value | 2.12192e-33 (rank : 29) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
NVL_MOUSE
|
||||||
| NC score | 0.917785 (rank : 26) | θ value | 3.37985e-39 (rank : 23) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
NVL_HUMAN
|
||||||
| NC score | 0.917151 (rank : 27) | θ value | 7.52953e-39 (rank : 24) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PRS7_HUMAN
|
||||||
| NC score | 0.916733 (rank : 28) | θ value | 4.00176e-32 (rank : 32) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P35998 | Gene names | PSMC2, MSS1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS7_MOUSE
|
||||||
| NC score | 0.916724 (rank : 29) | θ value | 4.00176e-32 (rank : 33) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P46471, O08531, Q3TGX5, Q9DBA1 | Gene names | Psmc2, Mss1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS6B_HUMAN
|
||||||
| NC score | 0.915946 (rank : 30) | θ value | 1.16434e-31 (rank : 35) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P43686, Q96FV5, Q9UBM3, Q9UEX3 | Gene names | PSMC4, TBP7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7). | |||||
|
PRS6B_MOUSE
|
||||||
| NC score | 0.914773 (rank : 31) | θ value | 5.77852e-31 (rank : 37) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P54775 | Gene names | Psmc4, Tbp7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7) (CIP21). | |||||
|
PEX6_HUMAN
|
||||||
| NC score | 0.913518 (rank : 32) | θ value | 2.67802e-36 (rank : 28) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PEX6_MOUSE
|
||||||
| NC score | 0.913375 (rank : 33) | θ value | 5.39604e-37 (rank : 27) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
YMEL1_MOUSE
|
||||||
| NC score | 0.901291 (rank : 34) | θ value | 2.12192e-33 (rank : 30) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
YMEL1_HUMAN
|
||||||
| NC score | 0.897769 (rank : 35) | θ value | 1.4233e-29 (rank : 40) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96TA2, Q9H1Q0, Q9UMR9 | Gene names | YME1L1, FTSH1, YME1L | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin- associated metalloprotease) (PAMP). | |||||
|
AFG31_MOUSE
|
||||||
| NC score | 0.896499 (rank : 36) | θ value | 1.37539e-32 (rank : 31) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q920A7 | Gene names | Afg3l1 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 1 (EC 3.4.24.-). | |||||
|
SPG7_HUMAN
|
||||||
| NC score | 0.896205 (rank : 37) | θ value | 3.17079e-29 (rank : 41) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UQ90, O75756, Q96IB0 | Gene names | SPG7, CAR, CMAR, PGN | |||
|
Domain Architecture |

|
|||||
| Description | Paraplegin (EC 3.4.24.-) (Spastic paraplegia protein 7). | |||||
|
AFG32_HUMAN
|
||||||
| NC score | 0.890656 (rank : 38) | θ value | 2.19584e-30 (rank : 38) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9Y4W6 | Gene names | AFG3L2 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-) (Paraplegin-like protein). | |||||
|
AFG32_MOUSE
|
||||||
| NC score | 0.890492 (rank : 39) | θ value | 2.86786e-30 (rank : 39) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8JZQ2 | Gene names | Afg3l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-). | |||||
|
NSF_HUMAN
|
||||||
| NC score | 0.876233 (rank : 40) | θ value | 3.75424e-22 (rank : 42) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P46459, Q9UKZ2 | Gene names | NSF | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein). | |||||
|
NSF_MOUSE
|
||||||
| NC score | 0.875738 (rank : 41) | θ value | 3.75424e-22 (rank : 43) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P46460 | Gene names | Nsf, Skd2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein) (SKD2 protein). | |||||
|
ATAD2_MOUSE
|
||||||
| NC score | 0.868947 (rank : 42) | θ value | 1.98606e-31 (rank : 36) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
ATAD2_HUMAN
|
||||||
| NC score | 0.851530 (rank : 43) | θ value | 8.91499e-32 (rank : 34) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
TRP13_MOUSE
|
||||||
| NC score | 0.713959 (rank : 44) | θ value | 1.80886e-08 (rank : 47) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q3UA06, Q3UQG6, Q9CWW8 | Gene names | Trip13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13). | |||||
|
TRP13_HUMAN
|
||||||
| NC score | 0.712445 (rank : 45) | θ value | 3.08544e-08 (rank : 48) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q15645, O15324 | Gene names | TRIP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13) (Human papillomavirus type 16 E1 protein- binding protein) (HPV16 E1 protein-binding protein) (16E1-BP). | |||||
|
ATD3A_HUMAN
|
||||||
| NC score | 0.638257 (rank : 46) | θ value | 1.80886e-08 (rank : 46) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9NVI7, Q8N275, Q96A50 | Gene names | ATAD3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3A. | |||||
|
ATD3B_HUMAN
|
||||||
| NC score | 0.622660 (rank : 47) | θ value | 2.79066e-09 (rank : 44) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
ATAD3_MOUSE
|
||||||
| NC score | 0.614020 (rank : 48) | θ value | 1.80886e-08 (rank : 45) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
BCS1_HUMAN
|
||||||
| NC score | 0.570372 (rank : 49) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y276, Q7Z2V7 | Gene names | BCS1L, BCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein) (H-BCS1). | |||||
|
BCS1_MOUSE
|
||||||
| NC score | 0.559813 (rank : 50) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9CZP5 | Gene names | Bcs1l | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein). | |||||
|
CLPX_HUMAN
|
||||||
| NC score | 0.425665 (rank : 51) | θ value | 0.00509761 (rank : 49) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
CLPX_MOUSE
|
||||||
| NC score | 0.422852 (rank : 52) | θ value | 0.00509761 (rank : 50) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JHS4, Q9WVD1 | Gene names | Clpx | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
LONM_HUMAN
|
||||||
| NC score | 0.413642 (rank : 53) | θ value | 0.125558 (rank : 56) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
RUVB1_HUMAN
|
||||||
| NC score | 0.324473 (rank : 54) | θ value | 0.0563607 (rank : 53) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y265, Q9BSX9 | Gene names | RUVBL1, NMP238, TIP49, TIP49A | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (Nuclear matrix protein 238) (NMP 238) (54 kDa erythrocyte cytosolic protein) (ECP-54) (TIP60-associated protein 54-alpha) (TAP54-alpha). | |||||
|
RUVB1_MOUSE
|
||||||
| NC score | 0.324434 (rank : 55) | θ value | 0.0563607 (rank : 54) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P60122, O35753 | Gene names | Ruvbl1, Tip49, Tip49a | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (DNA helicase p50). | |||||
|
WRIP1_HUMAN
|
||||||
| NC score | 0.230738 (rank : 56) | θ value | 0.47712 (rank : 63) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
WRIP1_MOUSE
|
||||||
| NC score | 0.229241 (rank : 57) | θ value | 0.62314 (rank : 67) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
THNSL_MOUSE
|
||||||
| NC score | 0.131743 (rank : 58) | θ value | 0.62314 (rank : 66) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BH55 | Gene names | Thnsl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
RFC1_MOUSE
|
||||||
| NC score | 0.131007 (rank : 59) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
RUVB2_HUMAN
|
||||||
| NC score | 0.123910 (rank : 60) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y230, Q6PK27, Q9Y361 | Gene names | RUVBL2, TIP48, TIP49B | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (48 kDa TATA box-binding protein-interacting protein) (48 kDa TBP-interacting protein) (TIP49b) (Repressing pontin 52) (Reptin 52) (51 kDa erythrocyte cytosolic protein) (ECP-51) (TIP60-associated protein 54-beta) (TAP54-beta). | |||||
|
RUVB2_MOUSE
|
||||||
| NC score | 0.120386 (rank : 61) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WTM5 | Gene names | Ruvbl2 | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (p47 protein). | |||||
|
RFC1_HUMAN
|
||||||
| NC score | 0.120103 (rank : 62) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
THNSL_HUMAN
|
||||||
| NC score | 0.119957 (rank : 63) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IYQ7, Q5VV21 | Gene names | THNSL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
RFC4_HUMAN
|
||||||
| NC score | 0.108797 (rank : 64) | θ value | 0.813845 (rank : 69) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35249, Q6FHX7 | Gene names | RFC4 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 4 (Replication factor C 37 kDa subunit) (RF-C 37 kDa subunit) (RFC37) (Activator 1 37 kDa subunit) (A1 37 kDa subunit). | |||||
|
KAD6_HUMAN
|
||||||
| NC score | 0.107701 (rank : 65) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
RFC4_MOUSE
|
||||||
| NC score | 0.097165 (rank : 66) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99J62 | Gene names | Rfc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Replication factor C subunit 4. | |||||
|
KAD6_MOUSE
|
||||||
| NC score | 0.082420 (rank : 67) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VCP8 | Gene names | Ak6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
IMUP_HUMAN
|
||||||
| NC score | 0.071806 (rank : 68) | θ value | 1.06291 (rank : 71) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9GZP8, Q96H58, Q9HCR4 | Gene names | IMUP, C19orf33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immortalization-up-regulated protein (Hepatocyte growth factor activator inhibitor type 2-related small protein) (HAI-2-related small protein) (H2RSP). | |||||
|
RFC5_HUMAN
|
||||||
| NC score | 0.060017 (rank : 69) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P40937 | Gene names | RFC5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RFC5_MOUSE
|
||||||
| NC score | 0.059264 (rank : 70) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9D0F6 | Gene names | Rfc5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
BANK1_MOUSE
|
||||||
| NC score | 0.038822 (rank : 71) | θ value | 0.125558 (rank : 55) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80VH0, Q3U178, Q8BRV6, Q8BRY8 | Gene names | Bank1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell scaffold protein with ankyrin repeats (Protein AVIEF). | |||||
|
BANK1_HUMAN
|
||||||
| NC score | 0.032564 (rank : 72) | θ value | 0.47712 (rank : 61) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NDB2, Q8N5K8, Q8NB56, Q8WYN5, Q9NWP2 | Gene names | BANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell scaffold protein with ankyrin repeats. | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.031967 (rank : 73) | θ value | 0.21417 (rank : 57) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
SYYM_MOUSE
|
||||||
| NC score | 0.030369 (rank : 74) | θ value | 0.21417 (rank : 58) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BYL4, Q6PAH7 | Gene names | Yars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.1) (Tyrosine--tRNA ligase) (TyrRS). | |||||
|
LIMA1_MOUSE
|
||||||
| NC score | 0.025879 (rank : 75) | θ value | 0.0113563 (rank : 51) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ERG0, Q9ERG1 | Gene names | Lima1, D15Ertd366e, Eplin | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm) (mEPLIN). | |||||
|
SYYM_HUMAN
|
||||||
| NC score | 0.023580 (rank : 76) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2Z4, Q9H817 | Gene names | YARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.1) (Tyrosine--tRNA ligase) (TyrRS). | |||||
|
THOC2_HUMAN
|
||||||
| NC score | 0.020650 (rank : 77) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
FOXM1_HUMAN
|
||||||
| NC score | 0.020471 (rank : 78) | θ value | 0.0252991 (rank : 52) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q08050, O43258, O43259, O43260, Q4ZGG7, Q9BRL2 | Gene names | FOXM1, FKHL16, HFH11, MPP2, WIN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead-related protein FKHL16) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/fork-head homolog 11) (HFH-11) (Winged helix factor from INS-1 cells) (M-phase phosphoprotein 2) (MPM-2 reactive phosphoprotein 2) (Transcription factor Trident). | |||||
|
TM131_MOUSE
|
||||||
| NC score | 0.018468 (rank : 79) | θ value | 1.06291 (rank : 73) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O70472 | Gene names | Tmem131, D1Bwg0491e, Rw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 131 (Protein RW1). | |||||
|
SMBP2_MOUSE
|
||||||
| NC score | 0.018451 (rank : 80) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P40694 | Gene names | Ighmbp2, Smbp-2, Smbp2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Cardiac transcription factor 1) (CATF1). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.016057 (rank : 81) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.014807 (rank : 82) | θ value | 0.62314 (rank : 65) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
RTKN_MOUSE
|
||||||
| NC score | 0.014788 (rank : 83) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C6B2, Q61192, Q8VIG7 | Gene names | Rtkn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rhotekin. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.014711 (rank : 84) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.014262 (rank : 85) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.013185 (rank : 86) | θ value | 0.62314 (rank : 64) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
SAHH3_HUMAN
|
||||||
| NC score | 0.012748 (rank : 87) | θ value | 0.813845 (rank : 70) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96HN2, O94917 | Gene names | KIAA0828 | |||
|
Domain Architecture |

|
|||||
| Description | Putative adenosylhomocysteinase 3 (EC 3.3.1.1) (S-adenosyl-L- homocysteine hydrolase 3) (AdoHcyase 3). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.012062 (rank : 88) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.011684 (rank : 89) | θ value | 1.06291 (rank : 72) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.011612 (rank : 90) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.009745 (rank : 91) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
SPTA3_HUMAN
|
||||||
| NC score | 0.009503 (rank : 92) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NHX4, Q86WX5, Q8N9Y6 | Gene names | SPATA3, TSARG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 3 (Testis spermatocyte apoptosis- related protein 1) (Testis and spermatogenesis cell-related protein 1). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.009474 (rank : 93) | θ value | 0.47712 (rank : 62) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
DYH5_HUMAN
|
||||||
| NC score | 0.009203 (rank : 94) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
SAHH3_MOUSE
|
||||||
| NC score | 0.008963 (rank : 95) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q68FL4, Q8BIH1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative adenosylhomocysteinase 3 (EC 3.3.1.1) (S-adenosyl-L- homocysteine hydrolase 3) (AdoHcyase 3). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.008704 (rank : 96) | θ value | 0.365318 (rank : 59) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
SF01_MOUSE
|
||||||
| NC score | 0.007650 (rank : 97) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64213, O08817, P70167, Q61454, Q921Z4 | Gene names | Sf1, Zfm1, Zfp162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (mZFM) (Zinc finger gene in MEN1 locus) (Mammalian branch point- binding protein mBBP) (BBP) (CW17). | |||||
|
FA44A_HUMAN
|
||||||
| NC score | 0.007605 (rank : 98) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
SF01_HUMAN
|
||||||
| NC score | 0.007575 (rank : 99) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.006995 (rank : 100) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
K0427_HUMAN
|
||||||
| NC score | 0.006312 (rank : 101) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43310, Q8IVD5 | Gene names | KIAA0427 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0427. | |||||
|
PP14D_HUMAN
|
||||||
| NC score | 0.006242 (rank : 102) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NXH3 | Gene names | PPP1R14D, GBPI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 14D (Gastrointestinal and brain-specific PP1-inhibitory protein 1) (GBPI-1). | |||||
|
TSH1_MOUSE
|
||||||
| NC score | 0.006147 (rank : 103) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.005742 (rank : 104) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
DEK_MOUSE
|
||||||
| NC score | 0.005488 (rank : 105) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
TBCD5_HUMAN
|
||||||
| NC score | 0.005420 (rank : 106) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92609 | Gene names | TBC1D5, KIAA0210 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 5. | |||||
|
CBLB_MOUSE
|
||||||
| NC score | 0.004919 (rank : 107) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3TTA7 | Gene names | Cblb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase CBL-B (EC 6.3.2.-) (Signal transduction protein CBL-B) (SH3-binding protein CBL-B) (Casitas B-lineage lymphoma proto-oncogene b). | |||||
|
TNNT2_HUMAN
|
||||||
| NC score | 0.004604 (rank : 108) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P45379, O60214, Q99596, Q99597, Q9UM96 | Gene names | TNNT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.004005 (rank : 109) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
TNNT2_MOUSE
|
||||||
| NC score | 0.003681 (rank : 110) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P50752, Q64360, Q64377 | Gene names | Tnnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
TGM5_MOUSE
|
||||||
| NC score | 0.003603 (rank : 111) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D7I9, Q3V1F9 | Gene names | Tgm5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-glutamine gamma-glutamyltransferase 5 (EC 2.3.2.13) (Transglutaminase-5) (TGase 5). | |||||
|
MAGI1_MOUSE
|
||||||
| NC score | 0.003376 (rank : 112) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
RU17_MOUSE
|
||||||
| NC score | 0.003017 (rank : 113) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62376, Q3UIW4 | Gene names | Snrp70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 SNRNP 70 kDa) (snRNP70). | |||||
|
RU17_HUMAN
|
||||||
| NC score | 0.002883 (rank : 114) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08621, P78493, P78494, Q15364, Q15686, Q15687, Q15689, Q99377, Q9UE45, Q9UE46, Q9UE47, Q9UE48, Q9UFQ6 | Gene names | SNRP70, RPU1, U1AP1 | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 snRNP 70 kDa) (snRNP70) (U1-70K). | |||||
|
PO6F2_MOUSE
|
||||||
| NC score | 0.002327 (rank : 115) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
CPXM2_HUMAN
|
||||||
| NC score | 0.002165 (rank : 116) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N436 | Gene names | CPXM2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
|
GIT1_HUMAN
|
||||||
| NC score | 0.000293 (rank : 117) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y2X7, Q86SS0, Q9BRJ4 | Gene names | GIT1 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase- interactor 1) (GRK-interacting protein 1) (Cool-associated and tyrosine-phosphorylated protein 1) (Cat-1). | |||||
|
BARX2_MOUSE
|
||||||
| NC score | -0.000016 (rank : 118) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08686 | Gene names | Barx2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein BarH-like 2. | |||||
|
DMD_HUMAN
|
||||||
| NC score | -0.000489 (rank : 119) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
UTRO_HUMAN
|
||||||
| NC score | -0.000556 (rank : 120) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | -0.003399 (rank : 121) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
PRP4B_HUMAN
|
||||||
| NC score | -0.004073 (rank : 122) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1030 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13523, Q8TDP2, Q96QT7, Q9UEE6 | Gene names | PRPF4B, KIAA0536, PRP4, PRP4H, PRP4K | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (PRP4 kinase). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | -0.004121 (rank : 123) | θ value | 0.365318 (rank : 60) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
CTCF_HUMAN
|
||||||
| NC score | -0.004244 (rank : 124) | θ value | 0.813845 (rank : 68) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49711 | Gene names | CTCF | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor CTCF (CCCTC-binding factor) (CTCFL paralog) (11-zinc finger protein). | |||||
|
M4K4_HUMAN
|
||||||
| NC score | -0.005119 (rank : 125) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
STK6_HUMAN
|
||||||
| NC score | -0.005186 (rank : 126) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14965, O60445, O75873, Q9BQD6, Q9UPG5 | Gene names | STK6, AIK, ARK1, AURA, BTAK, STK15 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 6 (EC 2.7.11.1) (Serine/threonine kinase 15) (Aurora/IPL1-related kinase 1) (Aurora-related kinase 1) (hARK1) (Aurora-A) (Breast-tumor-amplified kinase). | |||||