Please be patient as the page loads
|
SMBP2_MOUSE
|
||||||
| SwissProt Accessions | P40694 | Gene names | Ighmbp2, Smbp-2, Smbp2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Cardiac transcription factor 1) (CATF1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SMBP2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.979198 (rank : 2) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
SMBP2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P40694 | Gene names | Ighmbp2, Smbp-2, Smbp2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Cardiac transcription factor 1) (CATF1). | |||||
|
RENT1_HUMAN
|
||||||
| θ value | 1.15895e-47 (rank : 3) | NC score | 0.816595 (rank : 3) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92900, O00239, O43343, Q86Z25, Q92842 | Gene names | UPF1, KIAA0221, RENT1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (hUpf1). | |||||
|
RENT1_MOUSE
|
||||||
| θ value | 1.51363e-47 (rank : 4) | NC score | 0.816413 (rank : 4) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9EPU0, Q99PR4 | Gene names | Upf1, Rent1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (mUpf1). | |||||
|
SETX_HUMAN
|
||||||
| θ value | 9.52487e-34 (rank : 5) | NC score | 0.712045 (rank : 8) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
PR285_HUMAN
|
||||||
| θ value | 8.3442e-30 (rank : 6) | NC score | 0.714336 (rank : 5) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BYK8, Q3C2G2, Q4VXQ1, Q8TEF3, Q96ND3, Q9C094 | Gene names | PRIC285, KIAA1769 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal proliferator-activated receptor A-interacting complex 285 kDa protein (EC 3.6.1.-) (ATP-dependent helicase PRIC285) (PPAR-alpha- interacting complex protein 285) (PPAR-gamma DBD-interacting protein 1) (PDIP1). | |||||
|
M10L1_HUMAN
|
||||||
| θ value | 4.14116e-29 (rank : 7) | NC score | 0.713428 (rank : 7) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BXT6, Q5TGD5, Q8NBD4, Q9NXW3, Q9UFB3, Q9UGX9 | Gene names | MOV10L1 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase Mov10l1 (EC 3.6.1.-) (Moloney leukemia virus 10-like protein 1) (MOV10-like 1). | |||||
|
MOV10_MOUSE
|
||||||
| θ value | 2.35151e-24 (rank : 8) | NC score | 0.713571 (rank : 6) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P23249, Q9DC64 | Gene names | Mov10, Gb110 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase MOV-10 (EC 3.6.1.-) (Moloney leukemia virus 10 protein). | |||||
|
MOV10_HUMAN
|
||||||
| θ value | 1.5242e-23 (rank : 9) | NC score | 0.711502 (rank : 9) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HCE1, Q8TEF0, Q9BSY3, Q9BUJ9 | Gene names | MOV10, KIAA1631 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase MOV-10 (EC 3.6.1.-) (Moloney leukemia virus 10 protein). | |||||
|
M10L1_MOUSE
|
||||||
| θ value | 2.69047e-20 (rank : 10) | NC score | 0.695654 (rank : 10) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99MV5, Q7TPA9, Q8C3W0, Q924C2 | Gene names | Mov10l1, Champ | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase Mov10l1 (EC 3.6.1.-) (Moloney leukemia virus 10-like protein 1) (MOV10-like 1) (Cardiac helicase activated by MEF2 protein) (Cardiac-specific RNA helicase). | |||||
|
ZNFX1_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 11) | NC score | 0.476346 (rank : 14) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
HELZ_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 12) | NC score | 0.625801 (rank : 11) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
ZNFX1_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 13) | NC score | 0.472511 (rank : 15) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P2E3, Q9BQM7, Q9BQM8, Q9H8C1, Q9H9S2, Q9NUM1, Q9NWW1 | Gene names | ZNFX1, KIAA1404 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
AQR_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 14) | NC score | 0.483230 (rank : 13) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60306, Q2YDX9, Q6IRU8, Q6PIC8 | Gene names | AQR, KIAA0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius (Intron-binding protein of 160 kDa) (IBP160). | |||||
|
AQR_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 15) | NC score | 0.486195 (rank : 12) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CFQ3, P97871, Q3U9N1, Q3ULE8, Q80TX8 | Gene names | Aqr, Kiaa0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius. | |||||
|
NKRF_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 16) | NC score | 0.118952 (rank : 17) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BY02, Q8BJU5, Q9CRL2 | Gene names | Nkrf, Nrf | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF). | |||||
|
GLI1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 17) | NC score | 0.009089 (rank : 94) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08151, Q8TDN9 | Gene names | GLI1, GLI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene) (Oncogene GLI). | |||||
|
NKRF_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 18) | NC score | 0.110040 (rank : 18) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15226, Q9UJ91 | Gene names | NKRF, ITBA4, NRF | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF) (ITBA4 protein). | |||||
|
PEX6_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 19) | NC score | 0.048294 (rank : 32) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
DHX57_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 20) | NC score | 0.069688 (rank : 20) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX57_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 21) | NC score | 0.067839 (rank : 22) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 22) | NC score | 0.011767 (rank : 85) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
SC24D_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 23) | NC score | 0.041734 (rank : 40) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94855 | Gene names | SEC24D, KIAA0755 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24D (SEC24-related protein D). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 24) | NC score | 0.036130 (rank : 46) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
TRP13_HUMAN
|
||||||
| θ value | 0.125558 (rank : 25) | NC score | 0.074802 (rank : 19) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15645, O15324 | Gene names | TRIP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13) (Human papillomavirus type 16 E1 protein- binding protein) (HPV16 E1 protein-binding protein) (16E1-BP). | |||||
|
HORN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.036999 (rank : 44) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
TRP13_MOUSE
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.069472 (rank : 21) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3UA06, Q3UQG6, Q9CWW8 | Gene names | Trip13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13). | |||||
|
DHX36_MOUSE
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.054623 (rank : 29) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.037713 (rank : 43) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
ELOA2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.033950 (rank : 48) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IYF1, Q9P2V9 | Gene names | TCEB3B, TCEB3L | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A2 (Elongin-A2) (EloA2) (Transcription elongation factor B polypeptide 3B). | |||||
|
CCNT2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.028419 (rank : 55) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60583, O60582, Q5I1Y0 | Gene names | CCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T2 (CycT2). | |||||
|
DHX16_HUMAN
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.044471 (rank : 35) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
K1683_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.036726 (rank : 45) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.016910 (rank : 74) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.037737 (rank : 42) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
LMO6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.013218 (rank : 80) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43900, O76007 | Gene names | LMO6 | |||
|
Domain Architecture |
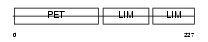
|
|||||
| Description | LIM domain only protein 6 (Triple LIM domain protein 6). | |||||
|
PHAR2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.032023 (rank : 49) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75167, Q68DM2 | Gene names | PHACTR2, C6orf56, KIAA0680 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 2. | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.034861 (rank : 47) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
PEG3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.003010 (rank : 109) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
PEX6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.041602 (rank : 41) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.028487 (rank : 54) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
CTND2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.015361 (rank : 75) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.004024 (rank : 106) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
SCN4A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.008658 (rank : 97) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SYN1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.027352 (rank : 57) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
DHX8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.043088 (rank : 38) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14562 | Gene names | DHX8, DDX8 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DHX8 (EC 3.6.1.-) (DEAH box protein 8) (RNA helicase HRH1). | |||||
|
DHX9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.048532 (rank : 31) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
TOP2A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.017543 (rank : 72) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11388, Q71UN1, Q9HB24, Q9HB25, Q9HB26, Q9UP44, Q9UQP9 | Gene names | TOP2A, TOP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
ABRA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.026221 (rank : 58) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N0Z2 | Gene names | ABRA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding Rho-activating protein (Striated muscle activator of Rho-dependent signaling) (STARS). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.021873 (rank : 63) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CLMN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.011032 (rank : 87) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.023854 (rank : 62) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
PEO1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.045748 (rank : 34) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96RR1, Q6MZX2, Q96RR0 | Gene names | PEO1, C10orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Twinkle protein, mitochondrial precursor (EC 3.6.1.-) (T7 gp4-like protein with intramitochondrial nucleoid localization) (T7-like mitochondrial DNA helicase) (Progressive external ophthalmoplegia 1 protein). | |||||
|
ABCF2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.011336 (rank : 86) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99LE6, Q3UA24, Q8C1S5, Q9JL48 | Gene names | Abcf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 2. | |||||
|
DHX37_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.042864 (rank : 39) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.020950 (rank : 65) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
KTNA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.018451 (rank : 71) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
|
MSH6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.014391 (rank : 77) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54276, O54710 | Gene names | Msh6, Gtmbp | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.024734 (rank : 60) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PEO1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.043944 (rank : 36) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CIW5, Q8K1Z1 | Gene names | Peo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Twinkle protein, mitochondrial precursor (EC 3.6.1.-) (T7 gp4-like protein with intramitochondrial nucleoid localization) (T7-like mitochondrial DNA helicase) (Progressive external ophthalmoplegia 1 protein homolog). | |||||
|
RA51B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.018939 (rank : 70) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15315, O60914, O75210, Q3Y4F8, Q86SY3, Q86SY4, Q86TR0, Q86U92, Q86U93, Q86U94, Q8N6H4, Q9UPL5 | Gene names | RAD51L1, RAD51B, REC2 | |||
|
Domain Architecture |
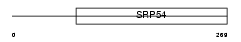
|
|||||
| Description | DNA repair protein RAD51 homolog 2 (R51H2) (RAD51-like protein 1) (Rad51B). | |||||
|
SYN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.024015 (rank : 61) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.020831 (rank : 66) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TMC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.015320 (rank : 76) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TDI7, Q5JXT0, Q5JXT1, Q6UWW4, Q6ZS41, Q8N9F3, Q9BYN2, Q9BYN3, Q9BYN4, Q9BYN5 | Gene names | TMC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
ASPX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.025890 (rank : 59) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26436 | Gene names | ACRV1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1). | |||||
|
MAST1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.003841 (rank : 107) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
PRP4B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.006880 (rank : 103) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
R3HD2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.020427 (rank : 68) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
RUSC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.017061 (rank : 73) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
DYHC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.028913 (rank : 53) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 733 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14204, Q6DKQ7, Q8WU28, Q92814, Q9Y4G5 | Gene names | DYNC1H1, DNCH1, DNECL, KIAA0325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
DYHC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.028988 (rank : 52) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JHU4 | Gene names | Dync1h1, Dnch1, Dnchc1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
K2C8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.009808 (rank : 91) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P05787, Q14099, Q14716, Q14717, Q53GJ0, Q6DHW5, Q6GMY0, Q96J60 | Gene names | KRT8, CYK8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.010795 (rank : 89) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RAI14_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.001906 (rank : 111) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
RN146_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.013951 (rank : 79) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CZW6, Q3TF93 | Gene names | Rnf146 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 146. | |||||
|
U2AFM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.014275 (rank : 78) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
VPS53_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.030727 (rank : 50) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5VIR6, Q8WYW3, Q9BRR2, Q9BY02, Q9NV25 | Gene names | VPS53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 53. | |||||
|
VPS53_MOUSE
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.030315 (rank : 51) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CCB4, Q8C7U0, Q8CIA1, Q9CX82, Q9D6T7 | Gene names | Vps53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 53. | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.010849 (rank : 88) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
CABP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.007132 (rank : 102) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CABP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.007827 (rank : 99) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VHC5 | Gene names | Cabp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CASP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.008861 (rank : 95) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CS029_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.019840 (rank : 69) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CS00 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf29 homolog. | |||||
|
FOXGA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.003483 (rank : 108) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55316 | Gene names | FOXG1A, FKHL2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein G1A (Forkhead-related protein FKHL2) (Transcription factor BF-2) (Brain factor 2) (BF2) (HFK2). | |||||
|
K2C8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.008726 (rank : 96) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 576 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P11679, Q3KQK5, Q3TGI1, Q3TJE1, Q3TKY7, Q61463, Q61518, Q61519 | Gene names | Krt8, Krt2-8 | |||
|
Domain Architecture |
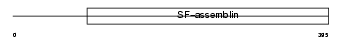
|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8) (Cytokeratin endo A). | |||||
|
OBSL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.007403 (rank : 101) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75147, Q96IW3 | Gene names | OBSL1, KIAA0657 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin-like protein 1 precursor. | |||||
|
R3HD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.020439 (rank : 67) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15032, Q8IW32 | Gene names | R3HDM1, KIAA0029, R3HDM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R3H domain-containing protein 1. | |||||
|
RAI1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.013143 (rank : 81) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
RBM16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.012912 (rank : 83) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
RFC5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.043405 (rank : 37) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P40937 | Gene names | RFC5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RFC5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.045790 (rank : 33) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9D0F6 | Gene names | Rfc5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
CM007_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.009745 (rank : 92) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K2Y0, Q9CYU1, Q9D1Y0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7 homolog. | |||||
|
CSMD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.002457 (rank : 110) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
DCAK3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.000150 (rank : 112) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BWQ5, Q1A748 | Gene names | Dcamkl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase DCAMKL3 (EC 2.7.11.1) (Doublecortin- like and CAM kinase-like 3) (CLICK-I and II-related) (CLr). | |||||
|
GPR98_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.005390 (rank : 105) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WXG9, O75171, Q9H0X5, Q9UL61 | Gene names | GPR98, KIAA0686, MASS1, VLGR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1 homolog) (Very large G-protein coupled receptor 1) (Usher syndrome type-2C protein). | |||||
|
LC7L2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.011811 (rank : 84) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TNC4, Q99LM5, Q99PC3 | Gene names | Luc7l2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2 (CGI-74 homolog). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.028162 (rank : 56) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.010610 (rank : 90) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.009481 (rank : 93) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
R51A1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.020986 (rank : 64) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
TEKT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.005682 (rank : 104) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q969V4 | Gene names | TEKT1 | |||
|
Domain Architecture |

|
|||||
| Description | Tektin-1. | |||||
|
U2AFL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.013061 (rank : 82) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.007881 (rank : 98) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
XYLT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.007779 (rank : 100) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPL0, Q5SUY1, Q8K060 | Gene names | Xylt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Xylosyltransferase 2 (EC 2.4.2.26) (Xylosyltransferase II) (Peptide O- xylosyltransferase 2). | |||||
|
CX048_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.182511 (rank : 16) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WUE5, Q9NWY8 | Gene names | CXorf48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CXorf48 (Tumor antigen BJ-HCC-20). | |||||
|
RRP44_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.061791 (rank : 23) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9Y2L1, Q5W0P7, Q5W0P8, Q658Z7, Q7Z481, Q8WWI2, Q9UG36 | Gene names | DIS3, KIAA1008, RRP44 | |||
|
Domain Architecture |

|
|||||
| Description | Exosome complex exonuclease RRP44 (EC 3.1.13.-) (Ribosomal RNA- processing protein 44) (DIS3 protein homolog). | |||||
|
SPR2B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.058842 (rank : 26) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70554, Q7TSB2 | Gene names | Sprr2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B. | |||||
|
SPR2D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.061056 (rank : 24) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70555 | Gene names | Sprr2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D. | |||||
|
SPR2E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.055057 (rank : 28) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22531, Q96RM2 | Gene names | SPRR2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E (SPR-2E) (Small proline-rich protein II) (SPR-II). | |||||
|
SPR2F_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.059029 (rank : 25) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96RM1 | Gene names | SPRR2F | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F (SPR-2F). | |||||
|
SPR2G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.058530 (rank : 27) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70558 | Gene names | Sprr2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G. | |||||
|
SPR2H_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.051631 (rank : 30) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70559 | Gene names | Sprr2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2H. | |||||
|
SMBP2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P40694 | Gene names | Ighmbp2, Smbp-2, Smbp2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Cardiac transcription factor 1) (CATF1). | |||||
|
SMBP2_HUMAN
|
||||||
| NC score | 0.979198 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
RENT1_HUMAN
|
||||||
| NC score | 0.816595 (rank : 3) | θ value | 1.15895e-47 (rank : 3) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92900, O00239, O43343, Q86Z25, Q92842 | Gene names | UPF1, KIAA0221, RENT1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (hUpf1). | |||||
|
RENT1_MOUSE
|
||||||
| NC score | 0.816413 (rank : 4) | θ value | 1.51363e-47 (rank : 4) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9EPU0, Q99PR4 | Gene names | Upf1, Rent1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (mUpf1). | |||||
|
PR285_HUMAN
|
||||||
| NC score | 0.714336 (rank : 5) | θ value | 8.3442e-30 (rank : 6) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BYK8, Q3C2G2, Q4VXQ1, Q8TEF3, Q96ND3, Q9C094 | Gene names | PRIC285, KIAA1769 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal proliferator-activated receptor A-interacting complex 285 kDa protein (EC 3.6.1.-) (ATP-dependent helicase PRIC285) (PPAR-alpha- interacting complex protein 285) (PPAR-gamma DBD-interacting protein 1) (PDIP1). | |||||
|
MOV10_MOUSE
|
||||||
| NC score | 0.713571 (rank : 6) | θ value | 2.35151e-24 (rank : 8) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P23249, Q9DC64 | Gene names | Mov10, Gb110 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase MOV-10 (EC 3.6.1.-) (Moloney leukemia virus 10 protein). | |||||
|
M10L1_HUMAN
|
||||||
| NC score | 0.713428 (rank : 7) | θ value | 4.14116e-29 (rank : 7) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BXT6, Q5TGD5, Q8NBD4, Q9NXW3, Q9UFB3, Q9UGX9 | Gene names | MOV10L1 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase Mov10l1 (EC 3.6.1.-) (Moloney leukemia virus 10-like protein 1) (MOV10-like 1). | |||||
|
SETX_HUMAN
|
||||||
| NC score | 0.712045 (rank : 8) | θ value | 9.52487e-34 (rank : 5) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
MOV10_HUMAN
|
||||||
| NC score | 0.711502 (rank : 9) | θ value | 1.5242e-23 (rank : 9) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HCE1, Q8TEF0, Q9BSY3, Q9BUJ9 | Gene names | MOV10, KIAA1631 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase MOV-10 (EC 3.6.1.-) (Moloney leukemia virus 10 protein). | |||||
|
M10L1_MOUSE
|
||||||
| NC score | 0.695654 (rank : 10) | θ value | 2.69047e-20 (rank : 10) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99MV5, Q7TPA9, Q8C3W0, Q924C2 | Gene names | Mov10l1, Champ | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase Mov10l1 (EC 3.6.1.-) (Moloney leukemia virus 10-like protein 1) (MOV10-like 1) (Cardiac helicase activated by MEF2 protein) (Cardiac-specific RNA helicase). | |||||
|
HELZ_HUMAN
|
||||||
| NC score | 0.625801 (rank : 11) | θ value | 4.16044e-13 (rank : 12) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
AQR_MOUSE
|
||||||
| NC score | 0.486195 (rank : 12) | θ value | 5.62301e-10 (rank : 15) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CFQ3, P97871, Q3U9N1, Q3ULE8, Q80TX8 | Gene names | Aqr, Kiaa0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius. | |||||
|
AQR_HUMAN
|
||||||
| NC score | 0.483230 (rank : 13) | θ value | 5.62301e-10 (rank : 14) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60306, Q2YDX9, Q6IRU8, Q6PIC8 | Gene names | AQR, KIAA0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius (Intron-binding protein of 160 kDa) (IBP160). | |||||
|
ZNFX1_MOUSE
|
||||||
| NC score | 0.476346 (rank : 14) | θ value | 2.20605e-14 (rank : 11) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
ZNFX1_HUMAN
|
||||||
| NC score | 0.472511 (rank : 15) | θ value | 5.43371e-13 (rank : 13) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P2E3, Q9BQM7, Q9BQM8, Q9H8C1, Q9H9S2, Q9NUM1, Q9NWW1 | Gene names | ZNFX1, KIAA1404 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
CX048_HUMAN
|
||||||
| NC score | 0.182511 (rank : 16) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WUE5, Q9NWY8 | Gene names | CXorf48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CXorf48 (Tumor antigen BJ-HCC-20). | |||||
|
NKRF_MOUSE
|
||||||
| NC score | 0.118952 (rank : 17) | θ value | 0.00228821 (rank : 16) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BY02, Q8BJU5, Q9CRL2 | Gene names | Nkrf, Nrf | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF). | |||||
|
NKRF_HUMAN
|
||||||
| NC score | 0.110040 (rank : 18) | θ value | 0.0252991 (rank : 18) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15226, Q9UJ91 | Gene names | NKRF, ITBA4, NRF | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF) (ITBA4 protein). | |||||
|
TRP13_HUMAN
|
||||||
| NC score | 0.074802 (rank : 19) | θ value | 0.125558 (rank : 25) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15645, O15324 | Gene names | TRIP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13) (Human papillomavirus type 16 E1 protein- binding protein) (HPV16 E1 protein-binding protein) (16E1-BP). | |||||
|
DHX57_MOUSE
|
||||||
| NC score | 0.069688 (rank : 20) | θ value | 0.0431538 (rank : 20) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
TRP13_MOUSE
|
||||||
| NC score | 0.069472 (rank : 21) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3UA06, Q3UQG6, Q9CWW8 | Gene names | Trip13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13). | |||||
|
DHX57_HUMAN
|
||||||
| NC score | 0.067839 (rank : 22) | θ value | 0.0736092 (rank : 21) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
RRP44_HUMAN
|
||||||
| NC score | 0.061791 (rank : 23) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9Y2L1, Q5W0P7, Q5W0P8, Q658Z7, Q7Z481, Q8WWI2, Q9UG36 | Gene names | DIS3, KIAA1008, RRP44 | |||
|
Domain Architecture |

|
|||||
| Description | Exosome complex exonuclease RRP44 (EC 3.1.13.-) (Ribosomal RNA- processing protein 44) (DIS3 protein homolog). | |||||
|
SPR2D_MOUSE
|
||||||
| NC score | 0.061056 (rank : 24) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70555 | Gene names | Sprr2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D. | |||||
|
SPR2F_HUMAN
|
||||||
| NC score | 0.059029 (rank : 25) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96RM1 | Gene names | SPRR2F | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F (SPR-2F). | |||||
|
SPR2B_MOUSE
|
||||||
| NC score | 0.058842 (rank : 26) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70554, Q7TSB2 | Gene names | Sprr2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B. | |||||
|
SPR2G_MOUSE
|
||||||
| NC score | 0.058530 (rank : 27) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70558 | Gene names | Sprr2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G. | |||||
|
SPR2E_HUMAN
|
||||||
| NC score | 0.055057 (rank : 28) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22531, Q96RM2 | Gene names | SPRR2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E (SPR-2E) (Small proline-rich protein II) (SPR-II). | |||||
|
DHX36_MOUSE
|
||||||
| NC score | 0.054623 (rank : 29) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
SPR2H_MOUSE
|
||||||
| NC score | 0.051631 (rank : 30) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70559 | Gene names | Sprr2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2H. | |||||
|
DHX9_HUMAN
|
||||||
| NC score | 0.048532 (rank : 31) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
PEX6_MOUSE
|
||||||
| NC score | 0.048294 (rank : 32) | θ value | 0.0252991 (rank : 19) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
RFC5_MOUSE
|
||||||
| NC score | 0.045790 (rank : 33) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9D0F6 | Gene names | Rfc5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
PEO1_HUMAN
|
||||||
| NC score | 0.045748 (rank : 34) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96RR1, Q6MZX2, Q96RR0 | Gene names | PEO1, C10orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Twinkle protein, mitochondrial precursor (EC 3.6.1.-) (T7 gp4-like protein with intramitochondrial nucleoid localization) (T7-like mitochondrial DNA helicase) (Progressive external ophthalmoplegia 1 protein). | |||||
|
DHX16_HUMAN
|
||||||
| NC score | 0.044471 (rank : 35) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
PEO1_MOUSE
|
||||||
| NC score | 0.043944 (rank : 36) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CIW5, Q8K1Z1 | Gene names | Peo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Twinkle protein, mitochondrial precursor (EC 3.6.1.-) (T7 gp4-like protein with intramitochondrial nucleoid localization) (T7-like mitochondrial DNA helicase) (Progressive external ophthalmoplegia 1 protein homolog). | |||||
|
RFC5_HUMAN
|
||||||
| NC score | 0.043405 (rank : 37) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P40937 | Gene names | RFC5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
DHX8_HUMAN
|
||||||
| NC score | 0.043088 (rank : 38) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14562 | Gene names | DHX8, DDX8 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DHX8 (EC 3.6.1.-) (DEAH box protein 8) (RNA helicase HRH1). | |||||
|
DHX37_HUMAN
|
||||||
| NC score | 0.042864 (rank : 39) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
SC24D_HUMAN
|
||||||
| NC score | 0.041734 (rank : 40) | θ value | 0.0736092 (rank : 23) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94855 | Gene names | SEC24D, KIAA0755 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24D (SEC24-related protein D). | |||||
|
PEX6_HUMAN
|
||||||
| NC score | 0.041602 (rank : 41) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.037737 (rank : 42) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.037713 (rank : 43) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
HORN_HUMAN
|
||||||
| NC score | 0.036999 (rank : 44) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
K1683_HUMAN
|
||||||
| NC score | 0.036726 (rank : 45) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.036130 (rank : 46) | θ value | 0.125558 (rank : 24) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.034861 (rank : 47) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
ELOA2_HUMAN
|
||||||
| NC score | 0.033950 (rank : 48) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IYF1, Q9P2V9 | Gene names | TCEB3B, TCEB3L | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A2 (Elongin-A2) (EloA2) (Transcription elongation factor B polypeptide 3B). | |||||
|
PHAR2_HUMAN
|
||||||
| NC score | 0.032023 (rank : 49) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75167, Q68DM2 | Gene names | PHACTR2, C6orf56, KIAA0680 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 2. | |||||
|
VPS53_HUMAN
|
||||||
| NC score | 0.030727 (rank : 50) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5VIR6, Q8WYW3, Q9BRR2, Q9BY02, Q9NV25 | Gene names | VPS53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 53. | |||||
|
VPS53_MOUSE
|
||||||
| NC score | 0.030315 (rank : 51) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CCB4, Q8C7U0, Q8CIA1, Q9CX82, Q9D6T7 | Gene names | Vps53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 53. | |||||
|
DYHC_MOUSE
|
||||||
| NC score | 0.028988 (rank : 52) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JHU4 | Gene names | Dync1h1, Dnch1, Dnchc1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
DYHC_HUMAN
|
||||||
| NC score | 0.028913 (rank : 53) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 733 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14204, Q6DKQ7, Q8WU28, Q92814, Q9Y4G5 | Gene names | DYNC1H1, DNCH1, DNECL, KIAA0325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.028487 (rank : 54) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
CCNT2_HUMAN
|
||||||
| NC score | 0.028419 (rank : 55) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60583, O60582, Q5I1Y0 | Gene names | CCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T2 (CycT2). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.028162 (rank : 56) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
SYN1_HUMAN
|
||||||
| NC score | 0.027352 (rank : 57) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
ABRA_HUMAN
|
||||||
| NC score | 0.026221 (rank : 58) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N0Z2 | Gene names | ABRA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding Rho-activating protein (Striated muscle activator of Rho-dependent signaling) (STARS). | |||||
|
ASPX_HUMAN
|
||||||
| NC score | 0.025890 (rank : 59) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26436 | Gene names | ACRV1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.024734 (rank : 60) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.024015 (rank : 61) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.023854 (rank : 62) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.021873 (rank : 63) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
R51A1_MOUSE
|
||||||
| NC score | 0.020986 (rank : 64) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.020950 (rank : 65) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.020831 (rank : 66) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
R3HD1_HUMAN
|
||||||
| NC score | 0.020439 (rank : 67) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15032, Q8IW32 | Gene names | R3HDM1, KIAA0029, R3HDM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R3H domain-containing protein 1. | |||||
|
R3HD2_MOUSE
|
||||||
| NC score | 0.020427 (rank : 68) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
CS029_MOUSE
|
||||||
| NC score | 0.019840 (rank : 69) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CS00 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf29 homolog. | |||||
|
RA51B_HUMAN
|
||||||
| NC score | 0.018939 (rank : 70) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15315, O60914, O75210, Q3Y4F8, Q86SY3, Q86SY4, Q86TR0, Q86U92, Q86U93, Q86U94, Q8N6H4, Q9UPL5 | Gene names | RAD51L1, RAD51B, REC2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA repair protein RAD51 homolog 2 (R51H2) (RAD51-like protein 1) (Rad51B). | |||||
|
KTNA1_MOUSE
|
||||||
| NC score | 0.018451 (rank : 71) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
|
TOP2A_HUMAN
|
||||||
| NC score | 0.017543 (rank : 72) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11388, Q71UN1, Q9HB24, Q9HB25, Q9HB26, Q9UP44, Q9UQP9 | Gene names | TOP2A, TOP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
RUSC2_MOUSE
|
||||||
| NC score | 0.017061 (rank : 73) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.016910 (rank : 74) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
CTND2_MOUSE
|
||||||
| NC score | 0.015361 (rank : 75) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
TMC2_HUMAN
|
||||||
| NC score | 0.015320 (rank : 76) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TDI7, Q5JXT0, Q5JXT1, Q6UWW4, Q6ZS41, Q8N9F3, Q9BYN2, Q9BYN3, Q9BYN4, Q9BYN5 | Gene names | TMC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
MSH6_MOUSE
|
||||||
| NC score | 0.014391 (rank : 77) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54276, O54710 | Gene names | Msh6, Gtmbp | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
U2AFM_HUMAN
|
||||||
| NC score | 0.014275 (rank : 78) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
RN146_MOUSE
|
||||||
| NC score | 0.013951 (rank : 79) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CZW6, Q3TF93 | Gene names | Rnf146 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 146. | |||||
|
LMO6_HUMAN
|
||||||
| NC score | 0.013218 (rank : 80) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43900, O76007 | Gene names | LMO6 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 6 (Triple LIM domain protein 6). | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.013143 (rank : 81) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
U2AFL_HUMAN
|
||||||
| NC score | 0.013061 (rank : 82) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
RBM16_HUMAN
|
||||||
| NC score | 0.012912 (rank : 83) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
LC7L2_MOUSE
|
||||||
| NC score | 0.011811 (rank : 84) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TNC4, Q99LM5, Q99PC3 | Gene names | Luc7l2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2 (CGI-74 homolog). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.011767 (rank : 85) | θ value | 0.0736092 (rank : 22) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
ABCF2_MOUSE
|
||||||
| NC score | 0.011336 (rank : 86) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99LE6, Q3UA24, Q8C1S5, Q9JL48 | Gene names | Abcf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 2. | |||||
|
CLMN_HUMAN
|
||||||
| NC score | 0.011032 (rank : 87) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.010849 (rank : 88) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.010795 (rank : 89) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.010610 (rank : 90) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
K2C8_HUMAN
|
||||||
| NC score | 0.009808 (rank : 91) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P05787, Q14099, Q14716, Q14717, Q53GJ0, Q6DHW5, Q6GMY0, Q96J60 | Gene names | KRT8, CYK8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8). | |||||
|
CM007_MOUSE
|
||||||
| NC score | 0.009745 (rank : 92) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K2Y0, Q9CYU1, Q9D1Y0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7 homolog. | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.009481 (rank : 93) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
GLI1_HUMAN
|
||||||
| NC score | 0.009089 (rank : 94) | θ value | 0.00869519 (rank : 17) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08151, Q8TDN9 | Gene names | GLI1, GLI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene) (Oncogene GLI). | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.008861 (rank : 95) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
K2C8_MOUSE
|
||||||
| NC score | 0.008726 (rank : 96) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 576 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P11679, Q3KQK5, Q3TGI1, Q3TJE1, Q3TKY7, Q61463, Q61518, Q61519 | Gene names | Krt8, Krt2-8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8) (Cytokeratin endo A). | |||||
|
SCN4A_HUMAN
|
||||||
| NC score | 0.008658 (rank : 97) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.007881 (rank : 98) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
CABP4_MOUSE
|
||||||
| NC score | 0.007827 (rank : 99) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VHC5 | Gene names | Cabp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
XYLT2_MOUSE
|
||||||
| NC score | 0.007779 (rank : 100) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPL0, Q5SUY1, Q8K060 | Gene names | Xylt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Xylosyltransferase 2 (EC 2.4.2.26) (Xylosyltransferase II) (Peptide O- xylosyltransferase 2). | |||||
|
OBSL1_HUMAN
|
||||||
| NC score | 0.007403 (rank : 101) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75147, Q96IW3 | Gene names | OBSL1, KIAA0657 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin-like protein 1 precursor. | |||||
|
CABP4_HUMAN
|
||||||
| NC score | 0.007132 (rank : 102) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
PRP4B_MOUSE
|
||||||
| NC score | 0.006880 (rank : 103) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
TEKT1_HUMAN
|
||||||
| NC score | 0.005682 (rank : 104) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q969V4 | Gene names | TEKT1 | |||
|
Domain Architecture |

|
|||||
| Description | Tektin-1. | |||||
|
GPR98_HUMAN
|
||||||
| NC score | 0.005390 (rank : 105) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WXG9, O75171, Q9H0X5, Q9UL61 | Gene names | GPR98, KIAA0686, MASS1, VLGR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1 homolog) (Very large G-protein coupled receptor 1) (Usher syndrome type-2C protein). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.004024 (rank : 106) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
MAST1_MOUSE
|
||||||
| NC score | 0.003841 (rank : 107) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
FOXGA_HUMAN
|
||||||
| NC score | 0.003483 (rank : 108) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55316 | Gene names | FOXG1A, FKHL2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein G1A (Forkhead-related protein FKHL2) (Transcription factor BF-2) (Brain factor 2) (BF2) (HFK2). | |||||
|
PEG3_MOUSE
|
||||||
| NC score | 0.003010 (rank : 109) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
CSMD2_HUMAN
|
||||||
| NC score | 0.002457 (rank : 110) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
RAI14_MOUSE
|
||||||
| NC score | 0.001906 (rank : 111) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
DCAK3_MOUSE
|
||||||
| NC score | 0.000150 (rank : 112) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BWQ5, Q1A748 | Gene names | Dcamkl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase DCAMKL3 (EC 2.7.11.1) (Doublecortin- like and CAM kinase-like 3) (CLICK-I and II-related) (CLr). | |||||