Please be patient as the page loads
|
TMC2_HUMAN
|
||||||
| SwissProt Accessions | Q8TDI7, Q5JXT0, Q5JXT1, Q6UWW4, Q6ZS41, Q8N9F3, Q9BYN2, Q9BYN3, Q9BYN4, Q9BYN5 | Gene names | TMC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TMC1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.947881 (rank : 3) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TDI8 | Gene names | TMC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 1. | |||||
|
TMC1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.958249 (rank : 2) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R4P5, Q9D435 | Gene names | Tmc1, Bth, dn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 1 (Beethoven protein) (Deafness protein). | |||||
|
TMC2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 149 | |
| SwissProt Accessions | Q8TDI7, Q5JXT0, Q5JXT1, Q6UWW4, Q6ZS41, Q8N9F3, Q9BYN2, Q9BYN3, Q9BYN4, Q9BYN5 | Gene names | TMC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
TMC2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.933720 (rank : 4) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8R4P4 | Gene names | Tmc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
TMC8_MOUSE
|
||||||
| θ value | 3.4976e-36 (rank : 5) | NC score | 0.765442 (rank : 5) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TN58, Q3TAL0, Q3UWI0, Q7TQ67 | Gene names | Tmc8, Ever2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane channel-like protein 8 (Epidermodysplasia verruciformis protein 2 homolog). | |||||
|
TMC8_HUMAN
|
||||||
| θ value | 9.52487e-34 (rank : 6) | NC score | 0.758635 (rank : 6) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IU68, Q8IWU7, Q8N358, Q8NF04 | Gene names | TMC8, EVER2, EVIN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane channel-like protein 8 (Epidermodysplasia verruciformis protein 2). | |||||
|
TMC6_MOUSE
|
||||||
| θ value | 7.30988e-26 (rank : 7) | NC score | 0.668851 (rank : 8) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TN60, Q78JR7, Q7TQ68, Q8BP52, Q8C6Z5, Q99J32 | Gene names | Tmc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane channel-like protein 6. | |||||
|
TMC6_HUMAN
|
||||||
| θ value | 3.62785e-25 (rank : 8) | NC score | 0.692990 (rank : 7) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z403, O43284, Q45VJ2, Q8IU98, Q8IWU8, Q8TEQ7, Q9HAG5 | Gene names | TMC6, EVER1, EVIN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane channel-like protein 6 (Epidermodysplasia verruciformis protein 1) (Protein LAK-4). | |||||
|
CING_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 9) | NC score | 0.034345 (rank : 44) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 10) | NC score | 0.060469 (rank : 16) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
SFRS6_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 11) | NC score | 0.054861 (rank : 22) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
RPTN_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 12) | NC score | 0.042818 (rank : 30) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
HAIR_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 13) | NC score | 0.041596 (rank : 31) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61645, Q80Y47 | Gene names | Hr | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 14) | NC score | 0.058082 (rank : 19) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 15) | NC score | 0.033815 (rank : 48) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 16) | NC score | 0.035649 (rank : 42) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PITX3_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 17) | NC score | 0.019469 (rank : 91) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35160 | Gene names | Pitx3 | |||
|
Domain Architecture |
|
|||||
| Description | Pituitary homeobox 3 (Homeobox protein PITX3). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 18) | NC score | 0.040053 (rank : 35) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
PITX3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 19) | NC score | 0.018903 (rank : 94) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75364 | Gene names | PITX3, PTX3 | |||
|
Domain Architecture |
|
|||||
| Description | Pituitary homeobox 3 (Homeobox protein PITX3). | |||||
|
MEFV_HUMAN
|
||||||
| θ value | 0.125558 (rank : 20) | NC score | 0.015818 (rank : 105) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 0.125558 (rank : 21) | NC score | 0.055225 (rank : 21) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
TLE4_MOUSE
|
||||||
| θ value | 0.125558 (rank : 22) | NC score | 0.024100 (rank : 74) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62441, Q9JKQ9 | Gene names | Tle4, Grg4 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4 (Groucho-related protein 4) (Grg- 4). | |||||
|
APC_MOUSE
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.052928 (rank : 23) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
TLE4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.023830 (rank : 75) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q04727, Q5T1Y2, Q9BZ07, Q9BZ08, Q9BZ09, Q9NSL3, Q9ULF9 | Gene names | TLE4, KIAA1261 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4. | |||||
|
CIR_MOUSE
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.065980 (rank : 11) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
EVL_MOUSE
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.033008 (rank : 52) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
NFRKB_HUMAN
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.039493 (rank : 37) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6P4R8, Q12869, Q15312, Q9H048 | Gene names | NFRKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.060073 (rank : 17) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
CI079_HUMAN
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.058281 (rank : 18) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
NUCB1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.039503 (rank : 36) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q02818, Q15838, Q7Z4J7, Q9BUR1 | Gene names | NUCB1, NUC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
PCGF2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.034326 (rank : 45) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35227 | Gene names | PCGF2, MEL18, RNF110, ZNF144 | |||
|
Domain Architecture |
|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144). | |||||
|
PCGF2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.033932 (rank : 47) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23798 | Gene names | Pcgf2, Mel-18, Mel18, Rnf110, Zfp144, Znf144 | |||
|
Domain Architecture |
|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144) (Zfp-144). | |||||
|
RNPS1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.049764 (rank : 24) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.049764 (rank : 25) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
XRN1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.026882 (rank : 66) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97789, O35651, P97790, Q3TE44, Q3TRE9, Q3UQL5 | Gene names | Xrn1, Dhm2, Exo, Sep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-3' exoribonuclease 1 (EC 3.1.11.-) (mXRN1) (Strand-exchange protein 1 homolog) (Protein Dhm2). | |||||
|
FNBP2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.014265 (rank : 112) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
SFRS4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.064767 (rank : 12) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
AKAP8_MOUSE
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.024693 (rank : 72) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBR0, Q9R0L8 | Gene names | Akap8, Akap95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
CTTB2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.015014 (rank : 110) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
GATA4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.019819 (rank : 90) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43694 | Gene names | GATA4 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
NEIL1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.034391 (rank : 43) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K4Q6, Q80V58, Q9CYT9 | Gene names | Neil1, Nei1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 1 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 1) (DNA glycosylase/AP lyase Neil1) (DNA-(apurinic or apyrimidinic site) lyase Neil1) (NEH1). | |||||
|
NUCB1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.039455 (rank : 38) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q02819, Q3TU42, Q9CWA1, Q9D8J4, Q9DBL3 | Gene names | Nucb1, Nuc, Nucb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
TTF1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.041093 (rank : 32) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62187, Q9JKK5 | Gene names | Ttf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (mTFF-I) (RNA polymerase I termination factor). | |||||
|
ZCH10_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.075281 (rank : 10) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.028976 (rank : 63) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.017969 (rank : 95) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
MILK2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.021822 (rank : 80) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.022781 (rank : 76) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
RL14_MOUSE
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.040528 (rank : 33) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CR57, Q9D8Q1 | Gene names | Rpl14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 60S ribosomal protein L14. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.061003 (rank : 14) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TGON2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.040246 (rank : 34) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
BRSK1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.000303 (rank : 148) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TDC3, Q5J5B5, Q8NDD0, Q8NDR4, Q8TDC2, Q96AV4, Q96JL4 | Gene names | BRSK1, KIAA1811, SAD1 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 1 (EC 2.7.11.1) (SAD1 kinase) (SAD1A). | |||||
|
CCD82_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.028826 (rank : 64) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N4S0, Q9H2Q5, Q9H5E3 | Gene names | CCDC82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 82. | |||||
|
K2027_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.031092 (rank : 58) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZVF9, Q8IVE4 | Gene names | KIAA2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
KIF4A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.011700 (rank : 122) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.036060 (rank : 40) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.032412 (rank : 53) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.019854 (rank : 89) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.062958 (rank : 13) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
CDSN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.034185 (rank : 46) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TPC1 | Gene names | Cdsn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor. | |||||
|
APC_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.047321 (rank : 27) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
IF16_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.021894 (rank : 79) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.030173 (rank : 59) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.007052 (rank : 135) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
SSR3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.000224 (rank : 149) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P32745 | Gene names | SSTR3 | |||
|
Domain Architecture |
|
|||||
| Description | Somatostatin receptor type 3 (SS3R) (SSR-28). | |||||
|
STMN2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.030085 (rank : 60) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q93045, O14952, Q6PK68 | Gene names | STMN2, SCG10, SCGN10 | |||
|
Domain Architecture |
|
|||||
| Description | Stathmin-2 (SCG10 protein) (Superior cervical ganglion-10 protein). | |||||
|
STMN2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.030085 (rank : 61) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P55821 | Gene names | Stmn2, Scg10, Scgn10, Stmb2 | |||
|
Domain Architecture |
|
|||||
| Description | Stathmin-2 (SCG10 protein) (Superior cervical ganglion-10 protein). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.021602 (rank : 82) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CD2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.024383 (rank : 73) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
CF055_MOUSE
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.022333 (rank : 78) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CR26 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | Protein C6orf55 homolog. | |||||
|
CFTR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.006900 (rank : 136) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P26361, Q63893, Q63894, Q9JKQ6 | Gene names | Cftr, Abcc7 | |||
|
Domain Architecture |

|
|||||
| Description | Cystic fibrosis transmembrane conductance regulator (CFTR) (cAMP- dependent chloride channel) (ATP-binding cassette transporter sub- family C member 7). | |||||
|
DPOLL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.015376 (rank : 106) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QXE2, Q9CTJ1 | Gene names | Poll, Polk | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase lambda (EC 2.7.7.7) (EC 4.2.99.-) (Pol Lambda) (DNA polymerase kappa). | |||||
|
FBX40_MOUSE
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.015280 (rank : 109) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P62932 | Gene names | Fbxo40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 40. | |||||
|
PININ_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.036534 (rank : 39) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
SEN34_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.021749 (rank : 81) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BMZ5, Q58EU2, Q99LV6, Q9CYW1 | Gene names | Tsen34, Leng5, Sen34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (Leukocyte receptor cluster member 5 homolog). | |||||
|
SMBP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.015320 (rank : 107) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P40694 | Gene names | Ighmbp2, Smbp-2, Smbp2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Cardiac transcription factor 1) (CATF1). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.058064 (rank : 20) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
WDR85_MOUSE
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.016677 (rank : 100) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CYU6, Q5RJV7, Q8BIT9 | Gene names | Wdr85 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 85. | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.014844 (rank : 111) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
BCLF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.033206 (rank : 50) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
BCLF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.035687 (rank : 41) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
ENAM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.018968 (rank : 93) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O55196 | Gene names | Enam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enamelin precursor. | |||||
|
FILA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.047457 (rank : 26) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.060689 (rank : 15) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.026049 (rank : 69) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
PCSK7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.008221 (rank : 130) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16549, Q9UL57 | Gene names | PCSK7, LPC, PC7, PC8, SPC7 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 7 precursor (EC 3.4.21.-) (Proprotein convertase PC7) (Subtilisin/kexin-like protease PC7) (Prohormone convertase PC7) (PC8) (hPC8) (Lymphoma proprotein convertase). | |||||
|
RAG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.025190 (rank : 71) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P15919 | Gene names | Rag1, Rag-1 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 1 (RAG-1). | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.021470 (rank : 83) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
SC5A1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.008494 (rank : 128) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C3K6 | Gene names | Slc5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/glucose cotransporter 1 (Na(+)/glucose cotransporter 1) (High affinity sodium-glucose cotransporter). | |||||
|
BAG4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.016809 (rank : 98) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95429, O95818 | Gene names | BAG4, SODD | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
CKAP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.015818 (rank : 104) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3V1H1, Q66LN2, Q8BSF0 | Gene names | Ckap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 2. | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.005738 (rank : 137) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
GBRA4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.003250 (rank : 143) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D6F4 | Gene names | Gabra4 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric-acid receptor subunit alpha-4 precursor (GABA(A) receptor subunit alpha-4). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.016231 (rank : 101) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
MARCS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.032071 (rank : 54) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.026460 (rank : 68) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PDE4C_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.004708 (rank : 141) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3UEI1, Q8K0P4 | Gene names | Pde4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17). | |||||
|
PININ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.031955 (rank : 56) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
PTPRA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.003189 (rank : 144) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18433, Q14513, Q7Z2I2, Q96TD9 | Gene names | PTPRA, PTPA | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha). | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.001464 (rank : 147) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
TNR6A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.032064 (rank : 55) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
TTC16_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.013705 (rank : 116) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C1F5 | Gene names | Ttc16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
TTF2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.013376 (rank : 117) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
VDP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.015287 (rank : 108) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60763 | Gene names | VDP | |||
|
Domain Architecture |

|
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
BRD3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.013219 (rank : 118) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.016713 (rank : 99) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.025450 (rank : 70) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DUT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.019908 (rank : 88) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P33316, O14785, Q16708, Q16860, Q96Q81 | Gene names | DUT | |||
|
Domain Architecture |

|
|||||
| Description | Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial precursor (EC 3.6.1.23) (dUTPase) (dUTP pyrophosphatase). | |||||
|
HNRPC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.019015 (rank : 92) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07910, P22628, Q53EX2, Q59FD3, Q5FWE8, Q86SF8, Q86U45, Q96HK7, Q96HM4, Q96IY5, Q9BTS3 | Gene names | HNRPC | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1 / hnRNP C2). | |||||
|
IFT81_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.022373 (rank : 77) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.020686 (rank : 87) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.043765 (rank : 29) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.010755 (rank : 125) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.017866 (rank : 96) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
PACS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.017062 (rank : 97) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6VY07, Q6PJY6, Q6PKB6, Q7Z590, Q7Z5W4, Q8N8K6, Q96MW0, Q9NW92, Q9ULP5 | Gene names | PACS1, KIAA1175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphofurin acidic cluster sorting protein 1 (PACS-1). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.026746 (rank : 67) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
RAG1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.021347 (rank : 84) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P15918, Q8NER2 | Gene names | RAG1, RNF74 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 1 (RAG-1) (RING finger protein 74). | |||||
|
RRS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.014011 (rank : 114) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15050, Q9BUX8 | Gene names | RRS1, KIAA0112, RRR | |||
|
Domain Architecture |
|
|||||
| Description | Ribosome biogenesis regulatory protein homolog. | |||||
|
TMPSD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.002620 (rank : 145) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
TSP50_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.002238 (rank : 146) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UI38 | Gene names | TSP50 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific protease-like protein 50 precursor. | |||||
|
ZCH10_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.033057 (rank : 51) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
ZN598_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.015853 (rank : 103) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86UK7, Q8IW49, Q8N3D9, Q96FG3, Q9H7J3 | Gene names | ZNF598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
ABI2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.008405 (rank : 129) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NYB9, Q13147, Q13249, Q13801, Q9BV70 | Gene names | ABI2, ARGBPIA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 2 (Abelson interactor 2) (Abi-2) (Abl-binding protein 3) (AblBP3) (Arg-binding protein 1) (ArgBP1). | |||||
|
ACD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.014202 (rank : 113) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96AP0, Q562H5, Q9H8F9 | Gene names | ACD, PIP1, PTOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adrenocortical dysplasia protein homolog (POT1 and TIN2-interacting protein). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.020723 (rank : 86) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.043910 (rank : 28) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CA065_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.021096 (rank : 85) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8N715, Q8N746, Q8NA93 | Gene names | C1orf65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf65. | |||||
|
CAC1C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.005310 (rank : 139) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1D_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.004701 (rank : 142) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CUL4B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.005420 (rank : 138) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13620, Q6PIE4, Q6UP07, Q7Z673, Q9BY37, Q9UEB7, Q9UED7 | Gene names | CUL4B, KIAA0695 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4B (CUL-4B). | |||||
|
EPN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.007983 (rank : 131) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
|
FMNL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.012750 (rank : 120) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
GATA4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.012820 (rank : 119) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08369, P97491, Q9QZK4 | Gene names | Gata4, Gata-4 | |||
|
Domain Architecture |
|
|||||
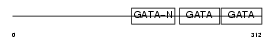
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.016032 (rank : 102) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
HASP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.010828 (rank : 124) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TF76, Q5U5K3, Q96MN1, Q9BXS7 | Gene names | GSG2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Haspin (EC 2.7.11.1) (Haploid germ cell-specific nuclear protein kinase) (H-Haspin) (Germ cell-specific gene 2 protein). | |||||
|
LBXCO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.010830 (rank : 123) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BX46, Q3UYA4, Q5W8I2, Q8C0T2 | Gene names | Lbxcor1, Corl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladybird homeobox corepressor 1 (Transcriptional corepressor Corl1). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.033637 (rank : 49) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MYLK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | -0.001442 (rank : 150) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15746, O95796, O95797, O95798, O95799, Q14844, Q16794, Q5MY99, Q5MYA0, Q7Z4J0, Q9C0L5, Q9UBG5, Q9UIT9 | Gene names | MYLK, MLCK | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.026889 (rank : 65) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NEBL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.007482 (rank : 132) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O76041, Q9UIC4 | Gene names | NEBL | |||
|
Domain Architecture |

|
|||||
| Description | Nebulette (Actin-binding Z-disk protein). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.012154 (rank : 121) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.029690 (rank : 62) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
SRPR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.009143 (rank : 127) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DBG7, Q8VC45, Q921H1 | Gene names | Srpr | |||
|
Domain Architecture |

|
|||||
| Description | Signal recognition particle receptor subunit alpha (SR-alpha) (Docking protein alpha) (DP-alpha). | |||||
|
TAB3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.010033 (rank : 126) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N5C8, Q6VQR0 | Gene names | MAP3K7IP3, TAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3) (NF-kappa-B-activating protein 1). | |||||
|
TGON1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.031140 (rank : 57) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.013747 (rank : 115) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
TWST2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.007275 (rank : 134) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WVJ9 | Gene names | TWIST2, DERMO1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TWST2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.007290 (rank : 133) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D030 | Gene names | Twist2, Dermo1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
UBP34_MOUSE
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.005159 (rank : 140) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
T2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.086290 (rank : 9) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q06666 | Gene names | Srst, T2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Octapeptide-repeat protein T2. | |||||
|
TMC2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 149 | |
| SwissProt Accessions | Q8TDI7, Q5JXT0, Q5JXT1, Q6UWW4, Q6ZS41, Q8N9F3, Q9BYN2, Q9BYN3, Q9BYN4, Q9BYN5 | Gene names | TMC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
TMC1_MOUSE
|
||||||
| NC score | 0.958249 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R4P5, Q9D435 | Gene names | Tmc1, Bth, dn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 1 (Beethoven protein) (Deafness protein). | |||||
|
TMC1_HUMAN
|
||||||
| NC score | 0.947881 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TDI8 | Gene names | TMC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 1. | |||||
|
TMC2_MOUSE
|
||||||
| NC score | 0.933720 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8R4P4 | Gene names | Tmc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
TMC8_MOUSE
|
||||||
| NC score | 0.765442 (rank : 5) | θ value | 3.4976e-36 (rank : 5) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TN58, Q3TAL0, Q3UWI0, Q7TQ67 | Gene names | Tmc8, Ever2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane channel-like protein 8 (Epidermodysplasia verruciformis protein 2 homolog). | |||||
|
TMC8_HUMAN
|
||||||
| NC score | 0.758635 (rank : 6) | θ value | 9.52487e-34 (rank : 6) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IU68, Q8IWU7, Q8N358, Q8NF04 | Gene names | TMC8, EVER2, EVIN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane channel-like protein 8 (Epidermodysplasia verruciformis protein 2). | |||||
|
TMC6_HUMAN
|
||||||
| NC score | 0.692990 (rank : 7) | θ value | 3.62785e-25 (rank : 8) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z403, O43284, Q45VJ2, Q8IU98, Q8IWU8, Q8TEQ7, Q9HAG5 | Gene names | TMC6, EVER1, EVIN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane channel-like protein 6 (Epidermodysplasia verruciformis protein 1) (Protein LAK-4). | |||||
|
TMC6_MOUSE
|
||||||
| NC score | 0.668851 (rank : 8) | θ value | 7.30988e-26 (rank : 7) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TN60, Q78JR7, Q7TQ68, Q8BP52, Q8C6Z5, Q99J32 | Gene names | Tmc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane channel-like protein 6. | |||||
|
T2_MOUSE
|
||||||
| NC score | 0.086290 (rank : 9) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q06666 | Gene names | Srst, T2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Octapeptide-repeat protein T2. | |||||
|
ZCH10_HUMAN
|
||||||
| NC score | 0.075281 (rank : 10) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.065980 (rank : 11) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
SFRS4_HUMAN
|
||||||
| NC score | 0.064767 (rank : 12) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.062958 (rank : 13) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.061003 (rank : 14) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.060689 (rank : 15) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.060469 (rank : 16) | θ value | 0.00390308 (rank : 10) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.060073 (rank : 17) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
CI079_HUMAN
|
||||||
| NC score | 0.058281 (rank : 18) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.058082 (rank : 19) | θ value | 0.0330416 (rank : 14) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.058064 (rank : 20) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.055225 (rank : 21) | θ value | 0.125558 (rank : 21) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
SFRS6_HUMAN
|
||||||
| NC score | 0.054861 (rank : 22) | θ value | 0.0113563 (rank : 11) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.052928 (rank : 23) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
RNPS1_HUMAN
|
||||||
| NC score | 0.049764 (rank : 24) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| NC score | 0.049764 (rank : 25) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.047457 (rank : 26) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.047321 (rank : 27) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.043910 (rank : 28) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.043765 (rank : 29) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.042818 (rank : 30) | θ value | 0.0193708 (rank : 12) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
HAIR_MOUSE
|
||||||
| NC score | 0.041596 (rank : 31) | θ value | 0.0252991 (rank : 13) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61645, Q80Y47 | Gene names | Hr | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
TTF1_MOUSE
|
||||||
| NC score | 0.041093 (rank : 32) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62187, Q9JKK5 | Gene names | Ttf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (mTFF-I) (RNA polymerase I termination factor). | |||||
|
RL14_MOUSE
|
||||||
| NC score | 0.040528 (rank : 33) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CR57, Q9D8Q1 | Gene names | Rpl14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 60S ribosomal protein L14. | |||||
|
TGON2_MOUSE
|
||||||
| NC score | 0.040246 (rank : 34) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.040053 (rank : 35) | θ value | 0.0736092 (rank : 18) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
NUCB1_HUMAN
|
||||||
| NC score | 0.039503 (rank : 36) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q02818, Q15838, Q7Z4J7, Q9BUR1 | Gene names | NUCB1, NUC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
NFRKB_HUMAN
|
||||||
| NC score | 0.039493 (rank : 37) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6P4R8, Q12869, Q15312, Q9H048 | Gene names | NFRKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
NUCB1_MOUSE
|
||||||
| NC score | 0.039455 (rank : 38) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q02819, Q3TU42, Q9CWA1, Q9D8J4, Q9DBL3 | Gene names | Nucb1, Nuc, Nucb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.036534 (rank : 39) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.036060 (rank : 40) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
BCLF1_MOUSE
|
||||||
| NC score | 0.035687 (rank : 41) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.035649 (rank : 42) | θ value | 0.0431538 (rank : 16) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NEIL1_MOUSE
|
||||||
| NC score | 0.034391 (rank : 43) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K4Q6, Q80V58, Q9CYT9 | Gene names | Neil1, Nei1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 1 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 1) (DNA glycosylase/AP lyase Neil1) (DNA-(apurinic or apyrimidinic site) lyase Neil1) (NEH1). | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.034345 (rank : 44) | θ value | 0.000786445 (rank : 9) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
PCGF2_HUMAN
|
||||||
| NC score | 0.034326 (rank : 45) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35227 | Gene names | PCGF2, MEL18, RNF110, ZNF144 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144). | |||||
|
CDSN_MOUSE
|
||||||
| NC score | 0.034185 (rank : 46) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TPC1 | Gene names | Cdsn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor. | |||||
|
PCGF2_MOUSE
|
||||||
| NC score | 0.033932 (rank : 47) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23798 | Gene names | Pcgf2, Mel-18, Mel18, Rnf110, Zfp144, Znf144 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144) (Zfp-144). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.033815 (rank : 48) | θ value | 0.0431538 (rank : 15) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.033637 (rank : 49) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
BCLF1_HUMAN
|
||||||
| NC score | 0.033206 (rank : 50) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
ZCH10_MOUSE
|
||||||
| NC score | 0.033057 (rank : 51) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
EVL_MOUSE
|
||||||
| NC score | 0.033008 (rank : 52) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.032412 (rank : 53) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.032071 (rank : 54) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
TNR6A_HUMAN
|
||||||
| NC score | 0.032064 (rank : 55) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.031955 (rank : 56) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
TGON1_MOUSE
|
||||||
| NC score | 0.031140 (rank : 57) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
K2027_HUMAN
|
||||||
| NC score | 0.031092 (rank : 58) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZVF9, Q8IVE4 | Gene names | KIAA2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.030173 (rank : 59) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
STMN2_HUMAN
|
||||||
| NC score | 0.030085 (rank : 60) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q93045, O14952, Q6PK68 | Gene names | STMN2, SCG10, SCGN10 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin-2 (SCG10 protein) (Superior cervical ganglion-10 protein). | |||||
|
STMN2_MOUSE
|
||||||
| NC score | 0.030085 (rank : 61) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P55821 | Gene names | Stmn2, Scg10, Scgn10, Stmb2 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin-2 (SCG10 protein) (Superior cervical ganglion-10 protein). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.029690 (rank : 62) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.028976 (rank : 63) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
CCD82_HUMAN
|
||||||
| NC score | 0.028826 (rank : 64) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N4S0, Q9H2Q5, Q9H5E3 | Gene names | CCDC82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 82. | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.026889 (rank : 65) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
XRN1_MOUSE
|
||||||
| NC score | 0.026882 (rank : 66) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97789, O35651, P97790, Q3TE44, Q3TRE9, Q3UQL5 | Gene names | Xrn1, Dhm2, Exo, Sep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-3' exoribonuclease 1 (EC 3.1.11.-) (mXRN1) (Strand-exchange protein 1 homolog) (Protein Dhm2). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.026746 (rank : 67) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.026460 (rank : 68) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.026049 (rank : 69) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.025450 (rank : 70) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
RAG1_MOUSE
|
||||||
| NC score | 0.025190 (rank : 71) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P15919 | Gene names | Rag1, Rag-1 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 1 (RAG-1). | |||||
|
AKAP8_MOUSE
|
||||||
| NC score | 0.024693 (rank : 72) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBR0, Q9R0L8 | Gene names | Akap8, Akap95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
CD2_HUMAN
|
||||||
| NC score | 0.024383 (rank : 73) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
TLE4_MOUSE
|
||||||
| NC score | 0.024100 (rank : 74) | θ value | 0.125558 (rank : 22) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62441, Q9JKQ9 | Gene names | Tle4, Grg4 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4 (Groucho-related protein 4) (Grg- 4). | |||||
|
TLE4_HUMAN
|
||||||
| NC score | 0.023830 (rank : 75) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q04727, Q5T1Y2, Q9BZ07, Q9BZ08, Q9BZ09, Q9NSL3, Q9ULF9 | Gene names | TLE4, KIAA1261 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4. | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.022781 (rank : 76) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
IFT81_HUMAN
|
||||||
| NC score | 0.022373 (rank : 77) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
CF055_MOUSE
|
||||||
| NC score | 0.022333 (rank : 78) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CR26 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C6orf55 homolog. | |||||
|
IF16_HUMAN
|
||||||
| NC score | 0.021894 (rank : 79) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
MILK2_HUMAN
|
||||||
| NC score | 0.021822 (rank : 80) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
SEN34_MOUSE
|
||||||
| NC score | 0.021749 (rank : 81) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BMZ5, Q58EU2, Q99LV6, Q9CYW1 | Gene names | Tsen34, Leng5, Sen34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (Leukocyte receptor cluster member 5 homolog). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.021602 (rank : 82) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.021470 (rank : 83) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
RAG1_HUMAN
|
||||||
| NC score | 0.021347 (rank : 84) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P15918, Q8NER2 | Gene names | RAG1, RNF74 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 1 (RAG-1) (RING finger protein 74). | |||||
|
CA065_HUMAN
|
||||||
| NC score | 0.021096 (rank : 85) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8N715, Q8N746, Q8NA93 | Gene names | C1orf65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf65. | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.020723 (rank : 86) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.020686 (rank : 87) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
DUT_HUMAN
|
||||||
| NC score | 0.019908 (rank : 88) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P33316, O14785, Q16708, Q16860, Q96Q81 | Gene names | DUT | |||
|
Domain Architecture |

|
|||||
| Description | Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial precursor (EC 3.6.1.23) (dUTPase) (dUTP pyrophosphatase). | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.019854 (rank : 89) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
GATA4_HUMAN
|
||||||
| NC score | 0.019819 (rank : 90) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43694 | Gene names | GATA4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
PITX3_MOUSE
|
||||||
| NC score | 0.019469 (rank : 91) | θ value | 0.0563607 (rank : 17) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35160 | Gene names | Pitx3 | |||
|
Domain Architecture |

|
|||||
| Description | Pituitary homeobox 3 (Homeobox protein PITX3). | |||||
|
HNRPC_HUMAN
|
||||||
| NC score | 0.019015 (rank : 92) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07910, P22628, Q53EX2, Q59FD3, Q5FWE8, Q86SF8, Q86U45, Q96HK7, Q96HM4, Q96IY5, Q9BTS3 | Gene names | HNRPC | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1 / hnRNP C2). | |||||
|
ENAM_MOUSE
|
||||||
| NC score | 0.018968 (rank : 93) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O55196 | Gene names | Enam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enamelin precursor. | |||||
|
PITX3_HUMAN
|
||||||
| NC score | 0.018903 (rank : 94) | θ value | 0.0961366 (rank : 19) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75364 | Gene names | PITX3, PTX3 | |||
|
Domain Architecture |

|
|||||
| Description | Pituitary homeobox 3 (Homeobox protein PITX3). | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | 0.017969 (rank : 95) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.017866 (rank : 96) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
PACS1_HUMAN
|
||||||
| NC score | 0.017062 (rank : 97) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6VY07, Q6PJY6, Q6PKB6, Q7Z590, Q7Z5W4, Q8N8K6, Q96MW0, Q9NW92, Q9ULP5 | Gene names | PACS1, KIAA1175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphofurin acidic cluster sorting protein 1 (PACS-1). | |||||
|
BAG4_HUMAN
|
||||||
| NC score | 0.016809 (rank : 98) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95429, O95818 | Gene names | BAG4, SODD | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.016713 (rank : 99) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
WDR85_MOUSE
|
||||||
| NC score | 0.016677 (rank : 100) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CYU6, Q5RJV7, Q8BIT9 | Gene names | Wdr85 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 85. | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.016231 (rank : 101) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.016032 (rank : 102) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
ZN598_HUMAN
|
||||||
| NC score | 0.015853 (rank : 103) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86UK7, Q8IW49, Q8N3D9, Q96FG3, Q9H7J3 | Gene names | ZNF598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
CKAP2_MOUSE
|
||||||
| NC score | 0.015818 (rank : 104) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3V1H1, Q66LN2, Q8BSF0 | Gene names | Ckap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 2. | |||||
|
MEFV_HUMAN
|
||||||
| NC score | 0.015818 (rank : 105) | θ value | 0.125558 (rank : 20) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
DPOLL_MOUSE
|
||||||
| NC score | 0.015376 (rank : 106) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QXE2, Q9CTJ1 | Gene names | Poll, Polk | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase lambda (EC 2.7.7.7) (EC 4.2.99.-) (Pol Lambda) (DNA polymerase kappa). | |||||
|
SMBP2_MOUSE
|
||||||
| NC score | 0.015320 (rank : 107) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P40694 | Gene names | Ighmbp2, Smbp-2, Smbp2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Cardiac transcription factor 1) (CATF1). | |||||
|
VDP_HUMAN
|
||||||
| NC score | 0.015287 (rank : 108) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60763 | Gene names | VDP | |||
|
Domain Architecture |

|
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
FBX40_MOUSE
|
||||||
| NC score | 0.015280 (rank : 109) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P62932 | Gene names | Fbxo40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 40. | |||||
|
CTTB2_HUMAN
|
||||||
| NC score | 0.015014 (rank : 110) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.014844 (rank : 111) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
FNBP2_HUMAN
|
||||||
| NC score | 0.014265 (rank : 112) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
ACD_HUMAN
|
||||||
| NC score | 0.014202 (rank : 113) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96AP0, Q562H5, Q9H8F9 | Gene names | ACD, PIP1, PTOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adrenocortical dysplasia protein homolog (POT1 and TIN2-interacting protein). | |||||
|
RRS1_HUMAN
|
||||||
| NC score | 0.014011 (rank : 114) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15050, Q9BUX8 | Gene names | RRS1, KIAA0112, RRR | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome biogenesis regulatory protein homolog. | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.013747 (rank : 115) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
TTC16_MOUSE
|
||||||
| NC score | 0.013705 (rank : 116) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C1F5 | Gene names | Ttc16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
TTF2_MOUSE
|
||||||
| NC score | 0.013376 (rank : 117) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
BRD3_MOUSE
|
||||||
| NC score | 0.013219 (rank : 118) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
GATA4_MOUSE
|
||||||
| NC score | 0.012820 (rank : 119) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08369, P97491, Q9QZK4 | Gene names | Gata4, Gata-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
FMNL_HUMAN
|
||||||
| NC score | 0.012750 (rank : 120) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.012154 (rank : 121) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
KIF4A_MOUSE
|
||||||
| NC score | 0.011700 (rank : 122) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
LBXCO_MOUSE
|
||||||
| NC score | 0.010830 (rank : 123) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BX46, Q3UYA4, Q5W8I2, Q8C0T2 | Gene names | Lbxcor1, Corl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladybird homeobox corepressor 1 (Transcriptional corepressor Corl1). | |||||
|
HASP_HUMAN
|
||||||
| NC score | 0.010828 (rank : 124) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TF76, Q5U5K3, Q96MN1, Q9BXS7 | Gene names | GSG2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Haspin (EC 2.7.11.1) (Haploid germ cell-specific nuclear protein kinase) (H-Haspin) (Germ cell-specific gene 2 protein). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.010755 (rank : 125) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
TAB3_HUMAN
|
||||||
| NC score | 0.010033 (rank : 126) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N5C8, Q6VQR0 | Gene names | MAP3K7IP3, TAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3) (NF-kappa-B-activating protein 1). | |||||
|
SRPR_MOUSE
|
||||||
| NC score | 0.009143 (rank : 127) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DBG7, Q8VC45, Q921H1 | Gene names | Srpr | |||
|
Domain Architecture |

|
|||||
| Description | Signal recognition particle receptor subunit alpha (SR-alpha) (Docking protein alpha) (DP-alpha). | |||||
|
SC5A1_MOUSE
|
||||||
| NC score | 0.008494 (rank : 128) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C3K6 | Gene names | Slc5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/glucose cotransporter 1 (Na(+)/glucose cotransporter 1) (High affinity sodium-glucose cotransporter). | |||||
|
ABI2_HUMAN
|
||||||
| NC score | 0.008405 (rank : 129) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NYB9, Q13147, Q13249, Q13801, Q9BV70 | Gene names | ABI2, ARGBPIA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 2 (Abelson interactor 2) (Abi-2) (Abl-binding protein 3) (AblBP3) (Arg-binding protein 1) (ArgBP1). | |||||
|
PCSK7_HUMAN
|
||||||
| NC score | 0.008221 (rank : 130) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16549, Q9UL57 | Gene names | PCSK7, LPC, PC7, PC8, SPC7 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 7 precursor (EC 3.4.21.-) (Proprotein convertase PC7) (Subtilisin/kexin-like protease PC7) (Prohormone convertase PC7) (PC8) (hPC8) (Lymphoma proprotein convertase). | |||||
|
EPN2_MOUSE
|
||||||
| NC score | 0.007983 (rank : 131) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
|
NEBL_HUMAN
|
||||||
| NC score | 0.007482 (rank : 132) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O76041, Q9UIC4 | Gene names | NEBL | |||
|
Domain Architecture |

|
|||||
| Description | Nebulette (Actin-binding Z-disk protein). | |||||
|
TWST2_MOUSE
|
||||||
| NC score | 0.007290 (rank : 133) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D030 | Gene names | Twist2, Dermo1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TWST2_HUMAN
|
||||||
| NC score | 0.007275 (rank : 134) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WVJ9 | Gene names | TWIST2, DERMO1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.007052 (rank : 135) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
CFTR_MOUSE
|
||||||
| NC score | 0.006900 (rank : 136) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P26361, Q63893, Q63894, Q9JKQ6 | Gene names | Cftr, Abcc7 | |||
|
Domain Architecture |

|
|||||
| Description | Cystic fibrosis transmembrane conductance regulator (CFTR) (cAMP- dependent chloride channel) (ATP-binding cassette transporter sub- family C member 7). | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.005738 (rank : 137) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
CUL4B_HUMAN
|
||||||
| NC score | 0.005420 (rank : 138) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13620, Q6PIE4, Q6UP07, Q7Z673, Q9BY37, Q9UEB7, Q9UED7 | Gene names | CUL4B, KIAA0695 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4B (CUL-4B). | |||||
|
CAC1C_MOUSE
|
||||||
| NC score | 0.005310 (rank : 139) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
UBP34_MOUSE
|
||||||
| NC score | 0.005159 (rank : 140) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
PDE4C_MOUSE
|
||||||
| NC score | 0.004708 (rank : 141) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3UEI1, Q8K0P4 | Gene names | Pde4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17). | |||||
|
CAC1D_HUMAN
|
||||||
| NC score | 0.004701 (rank : 142) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
GBRA4_MOUSE
|
||||||
| NC score | 0.003250 (rank : 143) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D6F4 | Gene names | Gabra4 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric-acid receptor subunit alpha-4 precursor (GABA(A) receptor subunit alpha-4). | |||||
|
PTPRA_HUMAN
|
||||||
| NC score | 0.003189 (rank : 144) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18433, Q14513, Q7Z2I2, Q96TD9 | Gene names | PTPRA, PTPA | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha). | |||||
|
TMPSD_MOUSE
|
||||||
| NC score | 0.002620 (rank : 145) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
TSP50_HUMAN
|
||||||
| NC score | 0.002238 (rank : 146) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UI38 | Gene names | TSP50 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific protease-like protein 50 precursor. | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.001464 (rank : 147) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
BRSK1_HUMAN
|
||||||
| NC score | 0.000303 (rank : 148) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TDC3, Q5J5B5, Q8NDD0, Q8NDR4, Q8TDC2, Q96AV4, Q96JL4 | Gene names | BRSK1, KIAA1811, SAD1 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 1 (EC 2.7.11.1) (SAD1 kinase) (SAD1A). | |||||
|
SSR3_HUMAN
|
||||||
| NC score | 0.000224 (rank : 149) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P32745 | Gene names | SSTR3 | |||
|
Domain Architecture |

|
|||||
| Description | Somatostatin receptor type 3 (SS3R) (SSR-28). | |||||
|
MYLK_HUMAN
|
||||||
| NC score | -0.001442 (rank : 150) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15746, O95796, O95797, O95798, O95799, Q14844, Q16794, Q5MY99, Q5MYA0, Q7Z4J0, Q9C0L5, Q9UBG5, Q9UIT9 | Gene names | MYLK, MLCK | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||