Please be patient as the page loads
|
CUL4B_HUMAN
|
||||||
| SwissProt Accessions | Q13620, Q6PIE4, Q6UP07, Q7Z673, Q9BY37, Q9UEB7, Q9UED7 | Gene names | CUL4B, KIAA0695 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4B (CUL-4B). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CUL4A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.993281 (rank : 3) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13619, O75834, Q589T6, Q5TC62, Q6UP08, Q9UP17 | Gene names | CUL4A | |||
|
Domain Architecture |
|
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
CUL4A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.993977 (rank : 2) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3TCH7, Q3THM3, Q91Z44 | Gene names | Cul4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
CUL4B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q13620, Q6PIE4, Q6UP07, Q7Z673, Q9BY37, Q9UEB7, Q9UED7 | Gene names | CUL4B, KIAA0695 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4B (CUL-4B). | |||||
|
CUL3_HUMAN
|
||||||
| θ value | 1.86219e-138 (rank : 4) | NC score | 0.956849 (rank : 5) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13618, O75415, Q569L3, Q9UBI8, Q9UET7 | Gene names | CUL3, KIAA0617 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
CUL3_MOUSE
|
||||||
| θ value | 1.86219e-138 (rank : 5) | NC score | 0.956938 (rank : 4) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JLV5 | Gene names | Cul3 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
CUL1_HUMAN
|
||||||
| θ value | 1.88829e-90 (rank : 6) | NC score | 0.925736 (rank : 6) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13616, O60719, Q8IYW1 | Gene names | CUL1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CUL1_MOUSE
|
||||||
| θ value | 1.88829e-90 (rank : 7) | NC score | 0.925727 (rank : 7) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WTX6, Q9WUI7 | Gene names | Cul1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CUL2_HUMAN
|
||||||
| θ value | 7.68416e-84 (rank : 8) | NC score | 0.921157 (rank : 8) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13617, O00200, Q9UNF9 | Gene names | CUL2 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-2 (CUL-2). | |||||
|
CUL2_MOUSE
|
||||||
| θ value | 3.56943e-81 (rank : 9) | NC score | 0.920468 (rank : 9) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D4H8, Q3TUR8 | Gene names | Cul2 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-2 (CUL-2). | |||||
|
CUL5_HUMAN
|
||||||
| θ value | 1.01059e-59 (rank : 10) | NC score | 0.889798 (rank : 11) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q93034, O14766, Q9BZC6 | Gene names | CUL5, VACM1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-5 (CUL-5) (Vasopressin-activated calcium-mobilizing receptor) (VACM-1). | |||||
|
CUL5_MOUSE
|
||||||
| θ value | 1.01059e-59 (rank : 11) | NC score | 0.890355 (rank : 10) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D5V5, Q8BMQ6, Q8BV53, Q8C098 | Gene names | Cul5 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-5 (CUL-5). | |||||
|
ANC2_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 12) | NC score | 0.451459 (rank : 12) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BZQ7, Q8R2Q1 | Gene names | Anapc2 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 2 (APC2) (Cyclosome subunit 2). | |||||
|
ANC2_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 13) | NC score | 0.449510 (rank : 13) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UJX6, Q5VSG1, Q96DG5, Q96GG4, Q9P2E1 | Gene names | ANAPC2, APC2, KIAA1406 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 2 (APC2) (Cyclosome subunit 2). | |||||
|
PARC_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 14) | NC score | 0.120236 (rank : 14) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
PARC_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 15) | NC score | 0.098267 (rank : 15) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80TT8, Q8BKL9, Q8CGC0, Q8R363 | Gene names | Parc, Kiaa0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein. | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 16) | NC score | 0.031276 (rank : 20) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 17) | NC score | 0.030431 (rank : 21) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
LAMA2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.029431 (rank : 23) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 0.125558 (rank : 19) | NC score | 0.032164 (rank : 19) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH14_HUMAN
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.026207 (rank : 28) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
RIF1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.025016 (rank : 34) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
ZN286_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | -0.000815 (rank : 83) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HBT8, Q96JF3 | Gene names | ZNF286, KIAA1874 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 286. | |||||
|
CE152_HUMAN
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.029101 (rank : 24) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
GBP4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.018935 (rank : 50) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96PP9, Q86T99 | Gene names | GBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate-binding protein 4 (GTP-binding protein 4) (Guanine nucleotide-binding protein 4) (GBP-4). | |||||
|
IQGA1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.024621 (rank : 35) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P46940 | Gene names | IQGAP1, KIAA0051 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1 (p195). | |||||
|
UACA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.019071 (rank : 49) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
CF152_HUMAN
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.025020 (rank : 33) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
HCDH_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.027183 (rank : 25) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16836, O00324, O00397, O00753 | Gene names | HADHSC, HAD, SCHAD | |||
|
Domain Architecture |
|
|||||
| Description | Short chain 3-hydroxyacyl-CoA dehydrogenase, mitochondrial precursor (EC 1.1.1.35) (HCDH) (Medium and short chain L-3-hydroxyacyl-coenzyme A dehydrogenase). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.026975 (rank : 26) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.010312 (rank : 63) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.010549 (rank : 62) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
CCD13_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.026224 (rank : 27) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
FYCO1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.021313 (rank : 45) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
HELI_MOUSE
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.000172 (rank : 82) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P81183 | Gene names | Ikzf2, Helios, Zfpn1a2, Znfn1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Helios (IKAROS family zinc finger protein 2). | |||||
|
IQGA1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.022797 (rank : 40) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JKF1 | Gene names | Iqgap1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1. | |||||
|
LAMA2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.025021 (rank : 32) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
LZTS1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.021676 (rank : 44) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P60853 | Gene names | Lzts1, Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/Esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
PARVG_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.014775 (rank : 52) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBI0, Q9BQX5, Q9NSG1 | Gene names | PARVG | |||
|
Domain Architecture |
|
|||||
| Description | Gamma-parvin. | |||||
|
PTN13_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.004360 (rank : 77) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.029809 (rank : 22) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CI093_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.022822 (rank : 39) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
DMD_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.017966 (rank : 51) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
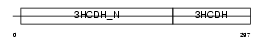
HCDH_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.024500 (rank : 36) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61425, Q3TF75, Q3THK8, Q3UFI0, Q8K149, Q925U9 | Gene names | Hadhsc, Hadh, Mschad, Schad | |||
|
Domain Architecture |
|
|||||
| Description | Short chain 3-hydroxyacyl-CoA dehydrogenase, mitochondrial precursor (EC 1.1.1.35) (HCDH) (Medium and short chain L-3-hydroxyacyl-coenzyme A dehydrogenase). | |||||
|
K0056_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.024399 (rank : 37) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42695 | Gene names | KIAA0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.000768 (rank : 81) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
K0056_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.022429 (rank : 43) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZQK0, Q9CS21 | Gene names | Kiaa0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
STXB4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.022558 (rank : 42) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 616 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6ZWJ1, Q8IVZ5 | Gene names | STXBP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
TMCC3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.014679 (rank : 53) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ULS5, Q8IWB2 | Gene names | TMCC3, KIAA1145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 3. | |||||
|
TPM2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.022621 (rank : 41) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 677 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P07951, P06468, Q13894, Q53FM4, Q5TCU4, Q5TCU7, Q9UH67 | Gene names | TPM2, TMSB | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
TPM2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.022971 (rank : 38) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P58774, P02560, P46901 | Gene names | Tpm2, Tpm-2 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.020594 (rank : 47) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
AHI1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.004263 (rank : 78) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 537 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N157, Q504T3, Q5TCP9, Q6P098, Q6PIT6, Q8NDX0, Q9H0H2 | Gene names | AHI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Jouberin (Abelson helper integration site 1 protein homolog) (AHI-1). | |||||
|
EXO1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.012848 (rank : 56) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZ11, Q3TLM4, Q923A5 | Gene names | Exo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (mExo1) (Exonuclease I). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.026080 (rank : 30) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.026132 (rank : 29) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.009583 (rank : 65) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
RTP3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.009883 (rank : 64) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
BTBDC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.013670 (rank : 54) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
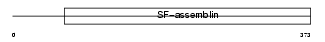
GFAP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.012066 (rank : 60) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P14136, Q53H98, Q5D055, Q6ZQS3, Q7Z5J6, Q7Z5J7, Q96KS4, Q96P18, Q9UFD0 | Gene names | GFAP | |||
|
Domain Architecture |
|
|||||
| Description | Glial fibrillary acidic protein, astrocyte (GFAP). | |||||
|
GP124_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.002624 (rank : 80) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91ZV8 | Gene names | Gpr124, Tem5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
IL6RA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.008981 (rank : 66) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22272 | Gene names | Il6ra, Il6r | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-6 receptor alpha chain precursor (IL-6R-alpha) (IL-6R 1) (CD126 antigen). | |||||
|
SIL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.013208 (rank : 55) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EPK6, Q91V34 | Gene names | Sil1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleotide exchange factor SIL1 precursor. | |||||
|
TPM4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.025395 (rank : 31) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6IRU2 | Gene names | Tpm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4). | |||||
|
VIME_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.012562 (rank : 59) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P08670, Q15867, Q15868, Q15869, Q6LER9, Q8N850, Q96ML2, Q9NTM3 | Gene names | VIM | |||
|
Domain Architecture |

|
|||||
| Description | Vimentin. | |||||
|
WAPL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.010759 (rank : 61) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q65Z40, Q6ZQE9 | Gene names | Wapal, Kiaa0261, Wapl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wings apart-like protein homolog (Dioxin-inducible factor 2) (DIF-2). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.004441 (rank : 76) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
DOCK4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.007766 (rank : 67) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.007655 (rank : 68) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
PAPOG_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.005879 (rank : 72) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PCL9, Q8BZC9 | Gene names | Papolg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) polymerase gamma (EC 2.7.7.19) (PAP gamma) (Polynucleotide adenylyltransferase gamma) (SRP RNA 3' adenylating enzyme). | |||||
|
TOM70_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.007523 (rank : 69) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94826 | Gene names | TOMM70A, KIAA0719 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
UBP10_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.006017 (rank : 71) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
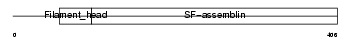
VIME_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.012597 (rank : 58) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P20152, O08704, Q8CCH1 | Gene names | Vim | |||
|
Domain Architecture |
|
|||||
| Description | Vimentin. | |||||
|
BCOR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.002682 (rank : 79) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
BICD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.020637 (rank : 46) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
BICD2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.019873 (rank : 48) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.004857 (rank : 74) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
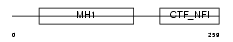
NFIB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.004491 (rank : 75) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00712, O00166, Q12858, Q96J45 | Gene names | NFIB | |||
|
Domain Architecture |
|
|||||
| Description | Nuclear factor 1 B-type (Nuclear factor 1/B) (NF1-B) (NFI-B) (NF-I/B) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
SACS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.012666 (rank : 57) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
TMC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.005420 (rank : 73) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TDI7, Q5JXT0, Q5JXT1, Q6UWW4, Q6ZS41, Q8N9F3, Q9BYN2, Q9BYN3, Q9BYN4, Q9BYN5 | Gene names | TMC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
TRIM8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.007105 (rank : 70) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BZR9, Q9C028 | Gene names | TRIM8, GERP, RNF27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
CJ046_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.079239 (rank : 18) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86Y37, Q5XPL7, Q8IY11, Q8N7S4 | Gene names | C10orf46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf46. | |||||
|
CJ046_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.080038 (rank : 17) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R0X2, Q3TE79, Q9CY95 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf46 homolog. | |||||
|
CUL7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.090433 (rank : 16) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14999, Q5T654 | Gene names | CUL7, KIAA0076 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-7 (CUL-7). | |||||
|
CUL4B_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q13620, Q6PIE4, Q6UP07, Q7Z673, Q9BY37, Q9UEB7, Q9UED7 | Gene names | CUL4B, KIAA0695 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4B (CUL-4B). | |||||
|
CUL4A_MOUSE
|
||||||
| NC score | 0.993977 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3TCH7, Q3THM3, Q91Z44 | Gene names | Cul4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
CUL4A_HUMAN
|
||||||
| NC score | 0.993281 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13619, O75834, Q589T6, Q5TC62, Q6UP08, Q9UP17 | Gene names | CUL4A | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
CUL3_MOUSE
|
||||||
| NC score | 0.956938 (rank : 4) | θ value | 1.86219e-138 (rank : 5) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JLV5 | Gene names | Cul3 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
CUL3_HUMAN
|
||||||
| NC score | 0.956849 (rank : 5) | θ value | 1.86219e-138 (rank : 4) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13618, O75415, Q569L3, Q9UBI8, Q9UET7 | Gene names | CUL3, KIAA0617 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
CUL1_HUMAN
|
||||||
| NC score | 0.925736 (rank : 6) | θ value | 1.88829e-90 (rank : 6) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13616, O60719, Q8IYW1 | Gene names | CUL1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CUL1_MOUSE
|
||||||
| NC score | 0.925727 (rank : 7) | θ value | 1.88829e-90 (rank : 7) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WTX6, Q9WUI7 | Gene names | Cul1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CUL2_HUMAN
|
||||||
| NC score | 0.921157 (rank : 8) | θ value | 7.68416e-84 (rank : 8) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13617, O00200, Q9UNF9 | Gene names | CUL2 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-2 (CUL-2). | |||||
|
CUL2_MOUSE
|
||||||
| NC score | 0.920468 (rank : 9) | θ value | 3.56943e-81 (rank : 9) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D4H8, Q3TUR8 | Gene names | Cul2 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-2 (CUL-2). | |||||
|
CUL5_MOUSE
|
||||||
| NC score | 0.890355 (rank : 10) | θ value | 1.01059e-59 (rank : 11) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D5V5, Q8BMQ6, Q8BV53, Q8C098 | Gene names | Cul5 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-5 (CUL-5). | |||||
|
CUL5_HUMAN
|
||||||
| NC score | 0.889798 (rank : 11) | θ value | 1.01059e-59 (rank : 10) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q93034, O14766, Q9BZC6 | Gene names | CUL5, VACM1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-5 (CUL-5) (Vasopressin-activated calcium-mobilizing receptor) (VACM-1). | |||||
|
ANC2_MOUSE
|
||||||
| NC score | 0.451459 (rank : 12) | θ value | 4.30538e-10 (rank : 12) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BZQ7, Q8R2Q1 | Gene names | Anapc2 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 2 (APC2) (Cyclosome subunit 2). | |||||
|
ANC2_HUMAN
|
||||||
| NC score | 0.449510 (rank : 13) | θ value | 9.59137e-10 (rank : 13) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UJX6, Q5VSG1, Q96DG5, Q96GG4, Q9P2E1 | Gene names | ANAPC2, APC2, KIAA1406 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 2 (APC2) (Cyclosome subunit 2). | |||||
|
PARC_HUMAN
|
||||||
| NC score | 0.120236 (rank : 14) | θ value | 4.1701e-05 (rank : 14) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
PARC_MOUSE
|
||||||
| NC score | 0.098267 (rank : 15) | θ value | 0.00228821 (rank : 15) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80TT8, Q8BKL9, Q8CGC0, Q8R363 | Gene names | Parc, Kiaa0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein. | |||||
|
CUL7_HUMAN
|
||||||
| NC score | 0.090433 (rank : 16) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14999, Q5T654 | Gene names | CUL7, KIAA0076 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-7 (CUL-7). | |||||
|
CJ046_MOUSE
|
||||||
| NC score | 0.080038 (rank : 17) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R0X2, Q3TE79, Q9CY95 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf46 homolog. | |||||
|
CJ046_HUMAN
|
||||||
| NC score | 0.079239 (rank : 18) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86Y37, Q5XPL7, Q8IY11, Q8N7S4 | Gene names | C10orf46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf46. | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.032164 (rank : 19) | θ value | 0.125558 (rank : 19) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.031276 (rank : 20) | θ value | 0.0113563 (rank : 16) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.030431 (rank : 21) | θ value | 0.0736092 (rank : 17) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.029809 (rank : 22) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
LAMA2_HUMAN
|
||||||
| NC score | 0.029431 (rank : 23) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.029101 (rank : 24) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
HCDH_HUMAN
|
||||||
| NC score | 0.027183 (rank : 25) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16836, O00324, O00397, O00753 | Gene names | HADHSC, HAD, SCHAD | |||
|
Domain Architecture |

|
|||||
| Description | Short chain 3-hydroxyacyl-CoA dehydrogenase, mitochondrial precursor (EC 1.1.1.35) (HCDH) (Medium and short chain L-3-hydroxyacyl-coenzyme A dehydrogenase). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.026975 (rank : 26) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
CCD13_HUMAN
|
||||||
| NC score | 0.026224 (rank : 27) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.026207 (rank : 28) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.026132 (rank : 29) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.026080 (rank : 30) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
TPM4_MOUSE
|
||||||
| NC score | 0.025395 (rank : 31) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6IRU2 | Gene names | Tpm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4). | |||||
|
LAMA2_MOUSE
|
||||||
| NC score | 0.025021 (rank : 32) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
CF152_HUMAN
|
||||||
| NC score | 0.025020 (rank : 33) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
RIF1_MOUSE
|
||||||
| NC score | 0.025016 (rank : 34) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
IQGA1_HUMAN
|
||||||
| NC score | 0.024621 (rank : 35) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P46940 | Gene names | IQGAP1, KIAA0051 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1 (p195). | |||||
|
HCDH_MOUSE
|
||||||
| NC score | 0.024500 (rank : 36) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61425, Q3TF75, Q3THK8, Q3UFI0, Q8K149, Q925U9 | Gene names | Hadhsc, Hadh, Mschad, Schad | |||
|
Domain Architecture |

|
|||||
| Description | Short chain 3-hydroxyacyl-CoA dehydrogenase, mitochondrial precursor (EC 1.1.1.35) (HCDH) (Medium and short chain L-3-hydroxyacyl-coenzyme A dehydrogenase). | |||||
|
K0056_HUMAN
|
||||||
| NC score | 0.024399 (rank : 37) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42695 | Gene names | KIAA0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
TPM2_MOUSE
|
||||||
| NC score | 0.022971 (rank : 38) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P58774, P02560, P46901 | Gene names | Tpm2, Tpm-2 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.022822 (rank : 39) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
IQGA1_MOUSE
|
||||||
| NC score | 0.022797 (rank : 40) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JKF1 | Gene names | Iqgap1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1. | |||||
|
TPM2_HUMAN
|
||||||
| NC score | 0.022621 (rank : 41) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 677 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P07951, P06468, Q13894, Q53FM4, Q5TCU4, Q5TCU7, Q9UH67 | Gene names | TPM2, TMSB | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
STXB4_HUMAN
|
||||||
| NC score | 0.022558 (rank : 42) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 616 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6ZWJ1, Q8IVZ5 | Gene names | STXBP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
K0056_MOUSE
|
||||||
| NC score | 0.022429 (rank : 43) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZQK0, Q9CS21 | Gene names | Kiaa0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
LZTS1_MOUSE
|
||||||
| NC score | 0.021676 (rank : 44) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P60853 | Gene names | Lzts1, Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/Esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
FYCO1_HUMAN
|
||||||
| NC score | 0.021313 (rank : 45) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
BICD1_HUMAN
|
||||||
| NC score | 0.020637 (rank : 46) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.020594 (rank : 47) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
BICD2_MOUSE
|
||||||
| NC score | 0.019873 (rank : 48) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
UACA_MOUSE
|
||||||
| NC score | 0.019071 (rank : 49) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
GBP4_HUMAN
|
||||||
| NC score | 0.018935 (rank : 50) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96PP9, Q86T99 | Gene names | GBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate-binding protein 4 (GTP-binding protein 4) (Guanine nucleotide-binding protein 4) (GBP-4). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.017966 (rank : 51) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
PARVG_HUMAN
|
||||||
| NC score | 0.014775 (rank : 52) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBI0, Q9BQX5, Q9NSG1 | Gene names | PARVG | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-parvin. | |||||
|
TMCC3_HUMAN
|
||||||
| NC score | 0.014679 (rank : 53) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ULS5, Q8IWB2 | Gene names | TMCC3, KIAA1145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 3. | |||||
|
BTBDC_HUMAN
|
||||||
| NC score | 0.013670 (rank : 54) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
SIL1_MOUSE
|
||||||
| NC score | 0.013208 (rank : 55) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EPK6, Q91V34 | Gene names | Sil1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleotide exchange factor SIL1 precursor. | |||||
|
EXO1_MOUSE
|
||||||
| NC score | 0.012848 (rank : 56) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZ11, Q3TLM4, Q923A5 | Gene names | Exo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (mExo1) (Exonuclease I). | |||||
|
SACS_MOUSE
|
||||||
| NC score | 0.012666 (rank : 57) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
VIME_MOUSE
|
||||||
| NC score | 0.012597 (rank : 58) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P20152, O08704, Q8CCH1 | Gene names | Vim | |||
|
Domain Architecture |

|
|||||
| Description | Vimentin. | |||||
|
VIME_HUMAN
|
||||||
| NC score | 0.012562 (rank : 59) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P08670, Q15867, Q15868, Q15869, Q6LER9, Q8N850, Q96ML2, Q9NTM3 | Gene names | VIM | |||
|
Domain Architecture |

|
|||||
| Description | Vimentin. | |||||
|
GFAP_HUMAN
|
||||||
| NC score | 0.012066 (rank : 60) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P14136, Q53H98, Q5D055, Q6ZQS3, Q7Z5J6, Q7Z5J7, Q96KS4, Q96P18, Q9UFD0 | Gene names | GFAP | |||
|
Domain Architecture |

|
|||||
| Description | Glial fibrillary acidic protein, astrocyte (GFAP). | |||||
|
WAPL_MOUSE
|
||||||
| NC score | 0.010759 (rank : 61) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q65Z40, Q6ZQE9 | Gene names | Wapal, Kiaa0261, Wapl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wings apart-like protein homolog (Dioxin-inducible factor 2) (DIF-2). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.010549 (rank : 62) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.010312 (rank : 63) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
RTP3_MOUSE
|
||||||
| NC score | 0.009883 (rank : 64) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | 0.009583 (rank : 65) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
IL6RA_MOUSE
|
||||||
| NC score | 0.008981 (rank : 66) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22272 | Gene names | Il6ra, Il6r | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-6 receptor alpha chain precursor (IL-6R-alpha) (IL-6R 1) (CD126 antigen). | |||||
|
DOCK4_HUMAN
|
||||||
| NC score | 0.007766 (rank : 67) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK4_MOUSE
|
||||||
| NC score | 0.007655 (rank : 68) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
TOM70_HUMAN
|
||||||
| NC score | 0.007523 (rank : 69) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94826 | Gene names | TOMM70A, KIAA0719 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
TRIM8_HUMAN
|
||||||
| NC score | 0.007105 (rank : 70) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BZR9, Q9C028 | Gene names | TRIM8, GERP, RNF27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
UBP10_MOUSE
|
||||||
| NC score | 0.006017 (rank : 71) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
PAPOG_MOUSE
|
||||||
| NC score | 0.005879 (rank : 72) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PCL9, Q8BZC9 | Gene names | Papolg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) polymerase gamma (EC 2.7.7.19) (PAP gamma) (Polynucleotide adenylyltransferase gamma) (SRP RNA 3' adenylating enzyme). | |||||
|
TMC2_HUMAN
|
||||||
| NC score | 0.005420 (rank : 73) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TDI7, Q5JXT0, Q5JXT1, Q6UWW4, Q6ZS41, Q8N9F3, Q9BYN2, Q9BYN3, Q9BYN4, Q9BYN5 | Gene names | TMC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.004857 (rank : 74) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NFIB_HUMAN
|
||||||
| NC score | 0.004491 (rank : 75) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00712, O00166, Q12858, Q96J45 | Gene names | NFIB | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 B-type (Nuclear factor 1/B) (NF1-B) (NFI-B) (NF-I/B) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.004441 (rank : 76) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
PTN13_HUMAN
|
||||||
| NC score | 0.004360 (rank : 77) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
AHI1_HUMAN
|
||||||
| NC score | 0.004263 (rank : 78) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 537 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N157, Q504T3, Q5TCP9, Q6P098, Q6PIT6, Q8NDX0, Q9H0H2 | Gene names | AHI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Jouberin (Abelson helper integration site 1 protein homolog) (AHI-1). | |||||
|
BCOR_MOUSE
|
||||||
| NC score | 0.002682 (rank : 79) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
GP124_MOUSE
|
||||||
| NC score | 0.002624 (rank : 80) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91ZV8 | Gene names | Gpr124, Tem5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.000768 (rank : 81) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
HELI_MOUSE
|
||||||
| NC score | 0.000172 (rank : 82) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P81183 | Gene names | Ikzf2, Helios, Zfpn1a2, Znfn1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Helios (IKAROS family zinc finger protein 2). | |||||
|
ZN286_HUMAN
|
||||||
| NC score | -0.000815 (rank : 83) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HBT8, Q96JF3 | Gene names | ZNF286, KIAA1874 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 286. | |||||