Please be patient as the page loads
|
SPAST_MOUSE
|
||||||
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SPAST_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.997290 (rank : 2) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q9UBP0, Q9UPR9 | Gene names | SPAST, KIAA1083, SPG4 | |||
|
Domain Architecture |

|
|||||
| Description | Spastin. | |||||
|
SPAST_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
FIGN_MOUSE
|
||||||
| θ value | 6.76295e-64 (rank : 3) | NC score | 0.939964 (rank : 13) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
KATL1_HUMAN
|
||||||
| θ value | 1.50663e-63 (rank : 4) | NC score | 0.962697 (rank : 4) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9BW62 | Gene names | KATNAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
FIGN_HUMAN
|
||||||
| θ value | 2.56992e-63 (rank : 5) | NC score | 0.935643 (rank : 14) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
KATL1_MOUSE
|
||||||
| θ value | 5.7252e-63 (rank : 6) | NC score | 0.963025 (rank : 3) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8K0T4 | Gene names | Katnal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
KTNA1_HUMAN
|
||||||
| θ value | 7.47731e-63 (rank : 7) | NC score | 0.961469 (rank : 5) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O75449, Q5TFA8, Q5TFA9, Q86VN2, Q9NU52 | Gene names | KATNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin). | |||||
|
KTNA1_MOUSE
|
||||||
| θ value | 6.32992e-62 (rank : 8) | NC score | 0.959766 (rank : 6) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
|
VPS4B_HUMAN
|
||||||
| θ value | 8.26713e-62 (rank : 9) | NC score | 0.957480 (rank : 9) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4B_MOUSE
|
||||||
| θ value | 1.41016e-61 (rank : 10) | NC score | 0.956956 (rank : 10) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4A_HUMAN
|
||||||
| θ value | 1.31987e-59 (rank : 11) | NC score | 0.953887 (rank : 11) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| θ value | 1.31987e-59 (rank : 12) | NC score | 0.953855 (rank : 12) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
ATAD1_HUMAN
|
||||||
| θ value | 2.3352e-48 (rank : 13) | NC score | 0.958906 (rank : 7) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8NBU5, Q6P4B9, Q8N3G1, Q8WYR9, Q969Y3 | Gene names | ATAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
ATAD1_MOUSE
|
||||||
| θ value | 2.3352e-48 (rank : 14) | NC score | 0.958906 (rank : 8) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9D5T0, Q3U8V2, Q9D7A4 | Gene names | Atad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
TERA_HUMAN
|
||||||
| θ value | 3.16345e-37 (rank : 15) | NC score | 0.920724 (rank : 17) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P55072, Q969G7 | Gene names | VCP | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
TERA_MOUSE
|
||||||
| θ value | 3.16345e-37 (rank : 16) | NC score | 0.920685 (rank : 18) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q01853 | Gene names | Vcp | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
NVL_HUMAN
|
||||||
| θ value | 2.67802e-36 (rank : 17) | NC score | 0.913628 (rank : 27) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
NVL_MOUSE
|
||||||
| θ value | 5.96599e-36 (rank : 18) | NC score | 0.913965 (rank : 26) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PRS8_HUMAN
|
||||||
| θ value | 1.12514e-34 (rank : 19) | NC score | 0.919217 (rank : 19) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P62195, O35051, O43208, P47210, P52915, P52916 | Gene names | PSMC5, SUG1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (Thyroid hormone receptor- interacting protein 1) (TRIP1). | |||||
|
PRS8_MOUSE
|
||||||
| θ value | 1.12514e-34 (rank : 20) | NC score | 0.919217 (rank : 20) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P62196, O35051, P47210, P52915, P52916, Q3UL51, Q9CWN5 | Gene names | Psmc5, Sug1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (mSUG1). | |||||
|
PRS10_HUMAN
|
||||||
| θ value | 2.50655e-34 (rank : 21) | NC score | 0.921638 (rank : 15) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P62333, P49719, Q6IBU3, Q92524 | Gene names | PSMC6, SUG2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS10_MOUSE
|
||||||
| θ value | 2.50655e-34 (rank : 22) | NC score | 0.921638 (rank : 16) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P62334, P49719, Q3TKK1, Q810A6, Q92524, Q9CXH9 | Gene names | Psmc6, Sug2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS4_HUMAN
|
||||||
| θ value | 2.12192e-33 (rank : 23) | NC score | 0.916139 (rank : 23) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P62191, P49014, Q03527, Q6IAW0, Q96AZ3 | Gene names | PSMC1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS4_MOUSE
|
||||||
| θ value | 2.12192e-33 (rank : 24) | NC score | 0.916139 (rank : 24) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P62192, P49014, Q03527, Q96AZ3 | Gene names | Psmc1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS6A_HUMAN
|
||||||
| θ value | 3.61944e-33 (rank : 25) | NC score | 0.917300 (rank : 22) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P17980, Q96HD3 | Gene names | PSMC3, TBP1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1) (Proteasome subunit P50). | |||||
|
PRS6A_MOUSE
|
||||||
| θ value | 4.72714e-33 (rank : 26) | NC score | 0.917412 (rank : 21) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
|
ATAD2_MOUSE
|
||||||
| θ value | 1.16434e-31 (rank : 27) | NC score | 0.866736 (rank : 41) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
ATAD2_HUMAN
|
||||||
| θ value | 1.52067e-31 (rank : 28) | NC score | 0.849238 (rank : 43) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
PEX6_MOUSE
|
||||||
| θ value | 3.3877e-31 (rank : 29) | NC score | 0.904334 (rank : 33) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PEX1_HUMAN
|
||||||
| θ value | 4.42448e-31 (rank : 30) | NC score | 0.914109 (rank : 25) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
PEX6_HUMAN
|
||||||
| θ value | 9.8567e-31 (rank : 31) | NC score | 0.904410 (rank : 32) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
AFG31_MOUSE
|
||||||
| θ value | 1.28732e-30 (rank : 32) | NC score | 0.893989 (rank : 37) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q920A7 | Gene names | Afg3l1 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 1 (EC 3.4.24.-). | |||||
|
AFG32_HUMAN
|
||||||
| θ value | 1.6813e-30 (rank : 33) | NC score | 0.889516 (rank : 39) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Y4W6 | Gene names | AFG3L2 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-) (Paraplegin-like protein). | |||||
|
AFG32_MOUSE
|
||||||
| θ value | 1.6813e-30 (rank : 34) | NC score | 0.889543 (rank : 38) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8JZQ2 | Gene names | Afg3l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-). | |||||
|
PRS6B_HUMAN
|
||||||
| θ value | 4.89182e-30 (rank : 35) | NC score | 0.912149 (rank : 28) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P43686, Q96FV5, Q9UBM3, Q9UEX3 | Gene names | PSMC4, TBP7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7). | |||||
|
PRS6B_MOUSE
|
||||||
| θ value | 2.42779e-29 (rank : 36) | NC score | 0.910988 (rank : 29) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P54775 | Gene names | Psmc4, Tbp7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7) (CIP21). | |||||
|
YMEL1_MOUSE
|
||||||
| θ value | 3.17079e-29 (rank : 37) | NC score | 0.897238 (rank : 34) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
PRS7_HUMAN
|
||||||
| θ value | 7.06379e-29 (rank : 38) | NC score | 0.910334 (rank : 30) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P35998 | Gene names | PSMC2, MSS1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS7_MOUSE
|
||||||
| θ value | 7.06379e-29 (rank : 39) | NC score | 0.910326 (rank : 31) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P46471, O08531, Q3TGX5, Q9DBA1 | Gene names | Psmc2, Mss1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
YMEL1_HUMAN
|
||||||
| θ value | 1.33217e-27 (rank : 40) | NC score | 0.894178 (rank : 36) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96TA2, Q9H1Q0, Q9UMR9 | Gene names | YME1L1, FTSH1, YME1L | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin- associated metalloprotease) (PAMP). | |||||
|
SPG7_HUMAN
|
||||||
| θ value | 1.12775e-26 (rank : 41) | NC score | 0.894784 (rank : 35) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UQ90, O75756, Q96IB0 | Gene names | SPG7, CAR, CMAR, PGN | |||
|
Domain Architecture |

|
|||||
| Description | Paraplegin (EC 3.4.24.-) (Spastic paraplegia protein 7). | |||||
|
NSF_HUMAN
|
||||||
| θ value | 2.27762e-19 (rank : 42) | NC score | 0.866928 (rank : 40) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P46459, Q9UKZ2 | Gene names | NSF | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein). | |||||
|
NSF_MOUSE
|
||||||
| θ value | 2.27762e-19 (rank : 43) | NC score | 0.866570 (rank : 42) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P46460 | Gene names | Nsf, Skd2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein) (SKD2 protein). | |||||
|
ATD3A_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 44) | NC score | 0.650376 (rank : 46) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9NVI7, Q8N275, Q96A50 | Gene names | ATAD3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3A. | |||||
|
ATD3B_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 45) | NC score | 0.631425 (rank : 47) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
ATAD3_MOUSE
|
||||||
| θ value | 1.63604e-09 (rank : 46) | NC score | 0.623862 (rank : 48) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
SPG20_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 47) | NC score | 0.152360 (rank : 59) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N0X7, O60349, Q86Y67, Q9H1T2, Q9H1T3 | Gene names | SPG20, KIAA0610, TAHCCP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spartin (Trans-activated by hepatitis C virus core protein 1). | |||||
|
SPG20_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 48) | NC score | 0.152692 (rank : 58) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R1X6, Q6ZQ87, Q8BJD3, Q8BM37, Q8BZ63 | Gene names | Spg20, Kiaa0610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spartin. | |||||
|
TRP13_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 49) | NC score | 0.701789 (rank : 45) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15645, O15324 | Gene names | TRIP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13) (Human papillomavirus type 16 E1 protein- binding protein) (HPV16 E1 protein-binding protein) (16E1-BP). | |||||
|
TRP13_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 50) | NC score | 0.705035 (rank : 44) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q3UA06, Q3UQG6, Q9CWW8 | Gene names | Trip13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13). | |||||
|
LONM_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 51) | NC score | 0.435653 (rank : 51) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
BCS1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 52) | NC score | 0.602644 (rank : 49) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y276, Q7Z2V7 | Gene names | BCS1L, BCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein) (H-BCS1). | |||||
|
CLPX_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 53) | NC score | 0.429934 (rank : 52) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
CLPX_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 54) | NC score | 0.426891 (rank : 53) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JHS4, Q9WVD1 | Gene names | Clpx | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
BCS1_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 55) | NC score | 0.587966 (rank : 50) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9CZP5 | Gene names | Bcs1l | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein). | |||||
|
RUVB1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 56) | NC score | 0.334037 (rank : 54) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y265, Q9BSX9 | Gene names | RUVBL1, NMP238, TIP49, TIP49A | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (Nuclear matrix protein 238) (NMP 238) (54 kDa erythrocyte cytosolic protein) (ECP-54) (TIP60-associated protein 54-alpha) (TAP54-alpha). | |||||
|
RUVB1_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 57) | NC score | 0.333997 (rank : 55) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P60122, O35753 | Gene names | Ruvbl1, Tip49, Tip49a | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (DNA helicase p50). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 0.163984 (rank : 58) | NC score | 0.034926 (rank : 73) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
SPN1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 59) | NC score | 0.038567 (rank : 72) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80W37 | Gene names | Snupn, Rnut1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Snurportin-1 (RNA U transporter 1). | |||||
|
MK15_HUMAN
|
||||||
| θ value | 0.365318 (rank : 60) | NC score | -0.002079 (rank : 140) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TD08, Q2TCF9, Q8N362 | Gene names | MAPK15, ERK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 15 (EC 2.7.11.24) (Extracellular signal-regulated kinase 8). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 61) | NC score | 0.020213 (rank : 78) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
WRIP1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 62) | NC score | 0.231533 (rank : 56) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.62314 (rank : 63) | NC score | 0.010411 (rank : 100) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
ST5_MOUSE
|
||||||
| θ value | 0.62314 (rank : 64) | NC score | 0.014855 (rank : 83) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q924W7, Q78H54, Q8K2P3, Q924W8 | Gene names | St5, Dennd2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (DENN domain-containing protein 2B). | |||||
|
WRIP1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 65) | NC score | 0.228754 (rank : 57) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.813845 (rank : 66) | NC score | 0.001782 (rank : 132) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.021710 (rank : 76) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ST5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.014568 (rank : 84) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78524, P78523, Q16492, Q7KYY2, Q7KZ12, Q8NE12, Q9BQQ6 | Gene names | ST5, DENND2B, HTS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (HeLa tumor suppression 1) (DENN domain-containing protein 2B). | |||||
|
BAT3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.018302 (rank : 80) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.012055 (rank : 89) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
FHOD1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.012340 (rank : 88) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P9Q4, Q8BMK2 | Gene names | Fhod1, Fhos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
ABCF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.011825 (rank : 90) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NE71, O14897, Q69YP6 | Gene names | ABCF1, ABC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1 (ATP-binding cassette 50) (TNF-alpha-stimulated ABC protein). | |||||
|
M3K15_HUMAN
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | -0.002084 (rank : 141) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 914 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6ZN16, Q5JPR4, Q6ZMV3 | Gene names | MAP3K15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 15 (EC 2.7.11.25) (MAPK/ERK kinase kinase 15) (MEK kinase 15) (MEKK 15). | |||||
|
M3K6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | -0.001959 (rank : 138) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95382 | Gene names | MAP3K6, MAPKKK6 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 6 (EC 2.7.11.25). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | -0.002236 (rank : 142) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MKL2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.010370 (rank : 101) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ULH7, Q68CT1, Q6UB16, Q86WW2, Q8N226 | Gene names | MKL2, KIAA1243, MRTFB | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B) (Megakaryoblastic leukemia 2). | |||||
|
ZDHC8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.006276 (rank : 114) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ULC8, Q6ICL1, Q6ZNF5, Q7Z6L9 | Gene names | ZDHHC8, KIAA1292, ZDHHCL1, ZNF378 | |||
|
Domain Architecture |
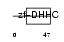
|
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8) (Zinc finger protein 378). | |||||
|
ZFHX2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.004100 (rank : 123) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
EP400_MOUSE
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.009996 (rank : 102) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
FOXL1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.002446 (rank : 128) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12952, Q9H242 | Gene names | FOXL1, FKHL11, FREAC7 | |||
|
Domain Architecture |
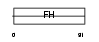
|
|||||
| Description | Forkhead box protein L1 (Forkhead-related protein FKHL11) (Forkhead- related transcription factor 7) (FREAC-7). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.007959 (rank : 107) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
NIP30_MOUSE
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.026730 (rank : 74) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WE2, Q8R0C9 | Gene names | Nip30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEFA-interacting nuclear protein NIP30. | |||||
|
NSMA2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.012845 (rank : 87) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NY59 | Gene names | SMPD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sphingomyelin phosphodiesterase 3 (EC 3.1.4.12) (Neutral sphingomyelinase 2) (Neutral sphingomyelinase II) (nSMase2) (nSMase- 2). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.011162 (rank : 94) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
YBOX2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.013052 (rank : 86) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2T7, Q8N4P0 | Gene names | YBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Y-box-binding protein 2 (Germ cell-specific Y-box-binding protein) (Contrin) (MSY2 homolog). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.007043 (rank : 113) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CND1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.010499 (rank : 98) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15021, Q8N6U3 | Gene names | NCAPD2, CAPD2, CNAP1, KIAA0159 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 1 (Non-SMC condensin I complex subunit D2) (Chromosome condensation-related SMC-associated protein 1) (Chromosome-associated protein D2) (hCAP-D2) (XCAP-D2 homolog). | |||||
|
LBXCO_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.009517 (rank : 103) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P84550 | Gene names | LBXCOR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladybird homeobox corepressor 1. | |||||
|
LETM1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.014122 (rank : 85) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z2I0, Q5PQC5, Q7TMK8, Q80ZX6, Q8CGJ3, Q8K5E5 | Gene names | Letm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper-EF-hand-containing transmembrane protein 1, mitochondrial precursor. | |||||
|
MCM5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.011812 (rank : 91) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49718 | Gene names | Mcm5, Cdc46, Mcmd5 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM5 (CDC46 homolog) (P1-CDC46). | |||||
|
OLIG1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.007924 (rank : 108) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TAK6, Q7RTS0 | Gene names | OLIG1 | |||
|
Domain Architecture |
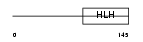
|
|||||
| Description | Oligodendrocyte transcription factor 1 (Oligo1). | |||||
|
RAD52_MOUSE
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.024463 (rank : 75) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43352 | Gene names | Rad52 | |||
|
Domain Architecture |
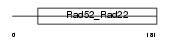
|
|||||
| Description | DNA repair protein RAD52 homolog. | |||||
|
RD23B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.011705 (rank : 93) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
RFC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.111310 (rank : 63) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
RFC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.121034 (rank : 62) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
RUVB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.131961 (rank : 60) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y230, Q6PK27, Q9Y361 | Gene names | RUVBL2, TIP48, TIP49B | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (48 kDa TATA box-binding protein-interacting protein) (48 kDa TBP-interacting protein) (TIP49b) (Repressing pontin 52) (Reptin 52) (51 kDa erythrocyte cytosolic protein) (ECP-51) (TIP60-associated protein 54-beta) (TAP54-beta). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.020865 (rank : 77) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SUHW3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | -0.000601 (rank : 135) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 697 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P3Y5, Q6ZPM2, Q8K145 | Gene names | Suhw3, Kiaa1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 3. | |||||
|
TACC3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.005093 (rank : 120) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.009055 (rank : 104) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.002947 (rank : 124) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.006163 (rank : 117) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.001848 (rank : 130) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
MCM5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.011051 (rank : 95) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P33992, O60785, Q14578, Q9BTJ4, Q9BWL8 | Gene names | MCM5, CDC46 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM5 (CDC46 homolog) (P1-CDC46). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.001209 (rank : 133) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.018906 (rank : 79) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.007566 (rank : 111) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CO6A2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.002078 (rank : 129) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P12110, Q13909, Q13910, Q13911, Q14049, Q16259, Q16597, Q9UML3 | Gene names | COL6A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(VI) chain precursor. | |||||
|
DIAP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.002696 (rank : 126) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
KAD6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.109915 (rank : 64) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
KLF12_MOUSE
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | -0.005302 (rank : 144) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35738 | Gene names | Klf12, Ap2rep | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 12 (Transcriptional repressor AP-2rep). | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.005566 (rank : 119) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
TTK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | -0.001729 (rank : 137) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35761 | Gene names | Ttk, Esk | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase TTK (EC 2.7.12.1) (ESK) (PYT). | |||||
|
ABCF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.011737 (rank : 92) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P542, Q6NV71 | Gene names | Abcf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1. | |||||
|
BRE1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.002835 (rank : 125) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
DYHC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.008982 (rank : 106) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 733 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14204, Q6DKQ7, Q8WU28, Q92814, Q9Y4G5 | Gene names | DYNC1H1, DNCH1, DNECL, KIAA0325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
DYHC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.009036 (rank : 105) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JHU4 | Gene names | Dync1h1, Dnch1, Dnchc1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
EHMT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | -0.001446 (rank : 136) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
GSHB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.007646 (rank : 110) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51855 | Gene names | Gss | |||
|
Domain Architecture |
|
|||||
| Description | Glutathione synthetase (EC 6.3.2.3) (Glutathione synthase) (GSH synthetase) (GSH-S). | |||||
|
LAP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | -0.002074 (rank : 139) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.001209 (rank : 134) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
PDE9A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.001796 (rank : 131) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O76083, O75490, O75491, O95225, Q86SF7, Q86SI6, Q86SJ3, Q86WN3, Q86WN4, Q86WN5, Q86WN6, Q86WN7, Q86WN8, Q86WN9, Q86WP0 | Gene names | PDE9A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PFD3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.017768 (rank : 82) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P61758, O55228, Q15765, Q5JT81, Q86X96 | Gene names | VBP1, PFDN3 | |||
|
Domain Architecture |
|
|||||
| Description | Prefoldin subunit 3 (Von Hippel-Lindau-binding protein 1) (VHL-binding protein 1) (VBP-1) (HIBBJ46). | |||||
|
PFD3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.017777 (rank : 81) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P61759, O55228, Q15765, Q3U6M9, Q9CPZ0 | Gene names | Vbp1, Pfdn3 | |||
|
Domain Architecture |

|
|||||
| Description | Prefoldin subunit 3 (Von Hippel-Lindau-binding protein 1) (VHL-binding protein 1) (VBP-1). | |||||
|
YBOX2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.010455 (rank : 99) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z2C8, Q5NCW8, Q5NCW9, Q9Z2C7 | Gene names | Ybx2, Msy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Y-box-binding protein 2 (Germ cell-specific Y-box-binding protein) (FRGY2 homolog). | |||||
|
CJ108_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.007093 (rank : 112) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N8Z3 | Gene names | C10orf108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative uncharacterized protein C10orf108. | |||||
|
CX039_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.006249 (rank : 115) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PEV8, Q8WVP6, Q96AV3 | Gene names | CXorf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein CXorf39. | |||||
|
EAN57_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.007762 (rank : 109) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43247, Q6ICF2, Q9Y4V8 | Gene names | EAN57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein EAN57. | |||||
|
IMMT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.004203 (rank : 122) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 494 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16891, Q14539, Q15092, Q68D41, Q69HW5, Q6IBL0, Q7Z3X1, Q8TAJ5, Q9P0V2 | Gene names | IMMT, HMP, PIG4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial inner membrane protein (Mitofilin) (p87/89) (Proliferation-inducing gene 4 protein). | |||||
|
KAD6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.087678 (rank : 68) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VCP8 | Gene names | Ak6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
KS6C1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.006224 (rank : 116) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96S38, Q8TDD3, Q9NSF4, Q9UL66 | Gene names | RPS6KC1, RPK118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase delta-1 (EC 2.7.11.1) (52 kDa ribosomal protein S6 kinase) (Ribosomal S6 kinase-like protein with two PSK domains 118 kDa protein) (SPHK1-binding protein). | |||||
|
LSD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.011007 (rank : 96) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6ZQ88, Q6PB53, Q8VEA1 | Gene names | Aof2, Kiaa0601, Lsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.010878 (rank : 97) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
RAVR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.002500 (rank : 127) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCJ3, Q6P141, Q9NPV7 | Gene names | RAVER2, KIAA1579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 2 (Protein raver-2). | |||||
|
RUVB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.125805 (rank : 61) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WTM5 | Gene names | Ruvbl2 | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (p47 protein). | |||||
|
TF3C2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.005793 (rank : 118) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WUA4, Q16632, Q9BWI7 | Gene names | GTF3C2, KIAA0011 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 2 (Transcription factor IIIC-subunit beta) (TF3C-beta) (TFIIIC 110 kDa subunit) (TFIIIC110). | |||||
|
THAP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.004643 (rank : 121) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WY91, Q53NU7, Q6GRN0, Q6IPJ3, Q9NW26, Q9Y325 | Gene names | THAP4 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 4. | |||||
|
TTK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | -0.002525 (rank : 143) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P33981 | Gene names | TTK, MPS1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase TTK (EC 2.7.12.1) (Phosphotyrosine picked threonine-protein kinase) (PYT). | |||||
|
RFC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.092456 (rank : 67) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35249, Q6FHX7 | Gene names | RFC4 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 4 (Replication factor C 37 kDa subunit) (RF-C 37 kDa subunit) (RFC37) (Activator 1 37 kDa subunit) (A1 37 kDa subunit). | |||||
|
RFC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.082292 (rank : 69) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99J62 | Gene names | Rfc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Replication factor C subunit 4. | |||||
|
RFC5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.054754 (rank : 70) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P40937 | Gene names | RFC5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RFC5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.054032 (rank : 71) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9D0F6 | Gene names | Rfc5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
THNSL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.097123 (rank : 66) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IYQ7, Q5VV21 | Gene names | THNSL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
THNSL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.104160 (rank : 65) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BH55 | Gene names | Thnsl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
SPAST_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
SPAST_HUMAN
|
||||||
| NC score | 0.997290 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q9UBP0, Q9UPR9 | Gene names | SPAST, KIAA1083, SPG4 | |||
|
Domain Architecture |

|
|||||
| Description | Spastin. | |||||
|
KATL1_MOUSE
|
||||||
| NC score | 0.963025 (rank : 3) | θ value | 5.7252e-63 (rank : 6) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8K0T4 | Gene names | Katnal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
KATL1_HUMAN
|
||||||
| NC score | 0.962697 (rank : 4) | θ value | 1.50663e-63 (rank : 4) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9BW62 | Gene names | KATNAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A-like 1 (EC 3.6.4.3) (Katanin p60 subunit A-like 1) (p60 katanin-like 1). | |||||
|
KTNA1_HUMAN
|
||||||
| NC score | 0.961469 (rank : 5) | θ value | 7.47731e-63 (rank : 7) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O75449, Q5TFA8, Q5TFA9, Q86VN2, Q9NU52 | Gene names | KATNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin). | |||||
|
KTNA1_MOUSE
|
||||||
| NC score | 0.959766 (rank : 6) | θ value | 6.32992e-62 (rank : 8) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9WV86 | Gene names | Katna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p60 ATPase-containing subunit A1 (EC 3.6.4.3) (Katanin p60 subunit A1) (p60 katanin) (Lipotransin). | |||||
|
ATAD1_HUMAN
|
||||||
| NC score | 0.958906 (rank : 7) | θ value | 2.3352e-48 (rank : 13) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8NBU5, Q6P4B9, Q8N3G1, Q8WYR9, Q969Y3 | Gene names | ATAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
ATAD1_MOUSE
|
||||||
| NC score | 0.958906 (rank : 8) | θ value | 2.3352e-48 (rank : 14) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9D5T0, Q3U8V2, Q9D7A4 | Gene names | Atad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 1. | |||||
|
VPS4B_HUMAN
|
||||||
| NC score | 0.957480 (rank : 9) | θ value | 8.26713e-62 (rank : 9) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4B_MOUSE
|
||||||
| NC score | 0.956956 (rank : 10) | θ value | 1.41016e-61 (rank : 10) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4A_HUMAN
|
||||||
| NC score | 0.953887 (rank : 11) | θ value | 1.31987e-59 (rank : 11) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4A_MOUSE
|
||||||
| NC score | 0.953855 (rank : 12) | θ value | 1.31987e-59 (rank : 12) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
FIGN_MOUSE
|
||||||
| NC score | 0.939964 (rank : 13) | θ value | 6.76295e-64 (rank : 3) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
FIGN_HUMAN
|
||||||
| NC score | 0.935643 (rank : 14) | θ value | 2.56992e-63 (rank : 5) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
PRS10_HUMAN
|
||||||
| NC score | 0.921638 (rank : 15) | θ value | 2.50655e-34 (rank : 21) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P62333, P49719, Q6IBU3, Q92524 | Gene names | PSMC6, SUG2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS10_MOUSE
|
||||||
| NC score | 0.921638 (rank : 16) | θ value | 2.50655e-34 (rank : 22) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P62334, P49719, Q3TKK1, Q810A6, Q92524, Q9CXH9 | Gene names | Psmc6, Sug2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
TERA_HUMAN
|
||||||
| NC score | 0.920724 (rank : 17) | θ value | 3.16345e-37 (rank : 15) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P55072, Q969G7 | Gene names | VCP | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
TERA_MOUSE
|
||||||
| NC score | 0.920685 (rank : 18) | θ value | 3.16345e-37 (rank : 16) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q01853 | Gene names | Vcp | |||
|
Domain Architecture |

|
|||||
| Description | Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin-containing protein) (VCP). | |||||
|
PRS8_HUMAN
|
||||||
| NC score | 0.919217 (rank : 19) | θ value | 1.12514e-34 (rank : 19) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P62195, O35051, O43208, P47210, P52915, P52916 | Gene names | PSMC5, SUG1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (Thyroid hormone receptor- interacting protein 1) (TRIP1). | |||||
|
PRS8_MOUSE
|
||||||
| NC score | 0.919217 (rank : 20) | θ value | 1.12514e-34 (rank : 20) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P62196, O35051, P47210, P52915, P52916, Q3UL51, Q9CWN5 | Gene names | Psmc5, Sug1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (mSUG1). | |||||
|
PRS6A_MOUSE
|
||||||
| NC score | 0.917412 (rank : 21) | θ value | 4.72714e-33 (rank : 26) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O88685 | Gene names | Psmc3, Tbp1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1). | |||||
|
PRS6A_HUMAN
|
||||||
| NC score | 0.917300 (rank : 22) | θ value | 3.61944e-33 (rank : 25) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P17980, Q96HD3 | Gene names | PSMC3, TBP1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6A (Proteasome 26S subunit ATPase 3) (TAT-binding protein 1) (TBP-1) (Proteasome subunit P50). | |||||
|
PRS4_HUMAN
|
||||||
| NC score | 0.916139 (rank : 23) | θ value | 2.12192e-33 (rank : 23) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P62191, P49014, Q03527, Q6IAW0, Q96AZ3 | Gene names | PSMC1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS4_MOUSE
|
||||||
| NC score | 0.916139 (rank : 24) | θ value | 2.12192e-33 (rank : 24) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P62192, P49014, Q03527, Q96AZ3 | Gene names | Psmc1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PEX1_HUMAN
|
||||||
| NC score | 0.914109 (rank : 25) | θ value | 4.42448e-31 (rank : 30) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
NVL_MOUSE
|
||||||
| NC score | 0.913965 (rank : 26) | θ value | 5.96599e-36 (rank : 18) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
NVL_HUMAN
|
||||||
| NC score | 0.913628 (rank : 27) | θ value | 2.67802e-36 (rank : 17) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PRS6B_HUMAN
|
||||||
| NC score | 0.912149 (rank : 28) | θ value | 4.89182e-30 (rank : 35) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P43686, Q96FV5, Q9UBM3, Q9UEX3 | Gene names | PSMC4, TBP7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7). | |||||
|
PRS6B_MOUSE
|
||||||
| NC score | 0.910988 (rank : 29) | θ value | 2.42779e-29 (rank : 36) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P54775 | Gene names | Psmc4, Tbp7 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 6B (Proteasome 26S subunit ATPase 4) (MIP224) (MB67-interacting protein) (TAT-binding protein 7) (TBP-7) (CIP21). | |||||
|
PRS7_HUMAN
|
||||||
| NC score | 0.910334 (rank : 30) | θ value | 7.06379e-29 (rank : 38) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P35998 | Gene names | PSMC2, MSS1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PRS7_MOUSE
|
||||||
| NC score | 0.910326 (rank : 31) | θ value | 7.06379e-29 (rank : 39) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P46471, O08531, Q3TGX5, Q9DBA1 | Gene names | Psmc2, Mss1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 7 (Proteasome 26S subunit ATPase 2) (Protein MSS1). | |||||
|
PEX6_HUMAN
|
||||||
| NC score | 0.904410 (rank : 32) | θ value | 9.8567e-31 (rank : 31) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PEX6_MOUSE
|
||||||
| NC score | 0.904334 (rank : 33) | θ value | 3.3877e-31 (rank : 29) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
YMEL1_MOUSE
|
||||||
| NC score | 0.897238 (rank : 34) | θ value | 3.17079e-29 (rank : 37) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
SPG7_HUMAN
|
||||||
| NC score | 0.894784 (rank : 35) | θ value | 1.12775e-26 (rank : 41) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UQ90, O75756, Q96IB0 | Gene names | SPG7, CAR, CMAR, PGN | |||
|
Domain Architecture |

|
|||||
| Description | Paraplegin (EC 3.4.24.-) (Spastic paraplegia protein 7). | |||||
|
YMEL1_HUMAN
|
||||||
| NC score | 0.894178 (rank : 36) | θ value | 1.33217e-27 (rank : 40) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96TA2, Q9H1Q0, Q9UMR9 | Gene names | YME1L1, FTSH1, YME1L | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin- associated metalloprotease) (PAMP). | |||||
|
AFG31_MOUSE
|
||||||
| NC score | 0.893989 (rank : 37) | θ value | 1.28732e-30 (rank : 32) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q920A7 | Gene names | Afg3l1 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 1 (EC 3.4.24.-). | |||||
|
AFG32_MOUSE
|
||||||
| NC score | 0.889543 (rank : 38) | θ value | 1.6813e-30 (rank : 34) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8JZQ2 | Gene names | Afg3l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-). | |||||
|
AFG32_HUMAN
|
||||||
| NC score | 0.889516 (rank : 39) | θ value | 1.6813e-30 (rank : 33) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Y4W6 | Gene names | AFG3L2 | |||
|
Domain Architecture |

|
|||||
| Description | AFG3-like protein 2 (EC 3.4.24.-) (Paraplegin-like protein). | |||||
|
NSF_HUMAN
|
||||||
| NC score | 0.866928 (rank : 40) | θ value | 2.27762e-19 (rank : 42) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P46459, Q9UKZ2 | Gene names | NSF | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein). | |||||
|
ATAD2_MOUSE
|
||||||
| NC score | 0.866736 (rank : 41) | θ value | 1.16434e-31 (rank : 27) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
NSF_MOUSE
|
||||||
| NC score | 0.866570 (rank : 42) | θ value | 2.27762e-19 (rank : 43) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P46460 | Gene names | Nsf, Skd2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-fusing ATPase (EC 3.6.4.6) (Vesicular-fusion protein NSF) (N- ethylmaleimide sensitive fusion protein) (NEM-sensitive fusion protein) (SKD2 protein). | |||||
|
ATAD2_HUMAN
|
||||||
| NC score | 0.849238 (rank : 43) | θ value | 1.52067e-31 (rank : 28) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
TRP13_MOUSE
|
||||||
| NC score | 0.705035 (rank : 44) | θ value | 9.92553e-07 (rank : 50) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q3UA06, Q3UQG6, Q9CWW8 | Gene names | Trip13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13). | |||||
|
TRP13_HUMAN
|
||||||
| NC score | 0.701789 (rank : 45) | θ value | 9.92553e-07 (rank : 49) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15645, O15324 | Gene names | TRIP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13) (Human papillomavirus type 16 E1 protein- binding protein) (HPV16 E1 protein-binding protein) (16E1-BP). | |||||
|
ATD3A_HUMAN
|
||||||
| NC score | 0.650376 (rank : 46) | θ value | 5.08577e-11 (rank : 44) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9NVI7, Q8N275, Q96A50 | Gene names | ATAD3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3A. | |||||
|
ATD3B_HUMAN
|
||||||
| NC score | 0.631425 (rank : 47) | θ value | 5.62301e-10 (rank : 45) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
ATAD3_MOUSE
|
||||||
| NC score | 0.623862 (rank : 48) | θ value | 1.63604e-09 (rank : 46) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
BCS1_HUMAN
|
||||||
| NC score | 0.602644 (rank : 49) | θ value | 0.00134147 (rank : 52) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y276, Q7Z2V7 | Gene names | BCS1L, BCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein) (H-BCS1). | |||||
|
BCS1_MOUSE
|
||||||
| NC score | 0.587966 (rank : 50) | θ value | 0.00665767 (rank : 55) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9CZP5 | Gene names | Bcs1l | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial chaperone BCS1 (BCS1-like protein). | |||||
|
LONM_HUMAN
|
||||||
| NC score | 0.435653 (rank : 51) | θ value | 0.00035302 (rank : 51) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
CLPX_HUMAN
|
||||||
| NC score | 0.429934 (rank : 52) | θ value | 0.00134147 (rank : 53) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
CLPX_MOUSE
|
||||||
| NC score | 0.426891 (rank : 53) | θ value | 0.00134147 (rank : 54) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JHS4, Q9WVD1 | Gene names | Clpx | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
RUVB1_HUMAN
|
||||||
| NC score | 0.334037 (rank : 54) | θ value | 0.00665767 (rank : 56) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y265, Q9BSX9 | Gene names | RUVBL1, NMP238, TIP49, TIP49A | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (Nuclear matrix protein 238) (NMP 238) (54 kDa erythrocyte cytosolic protein) (ECP-54) (TIP60-associated protein 54-alpha) (TAP54-alpha). | |||||
|
RUVB1_MOUSE
|
||||||
| NC score | 0.333997 (rank : 55) | θ value | 0.00665767 (rank : 57) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P60122, O35753 | Gene names | Ruvbl1, Tip49, Tip49a | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 1 (EC 3.6.1.-) (49 kDa TATA box-binding protein-interacting protein) (49 kDa TBP-interacting protein) (TIP49a) (Pontin 52) (DNA helicase p50). | |||||
|
WRIP1_HUMAN
|
||||||
| NC score | 0.231533 (rank : 56) | θ value | 0.365318 (rank : 62) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
WRIP1_MOUSE
|
||||||
| NC score | 0.228754 (rank : 57) | θ value | 0.62314 (rank : 65) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
SPG20_MOUSE
|
||||||
| NC score | 0.152692 (rank : 58) | θ value | 4.0297e-08 (rank : 48) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R1X6, Q6ZQ87, Q8BJD3, Q8BM37, Q8BZ63 | Gene names | Spg20, Kiaa0610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spartin. | |||||
|
SPG20_HUMAN
|
||||||
| NC score | 0.152360 (rank : 59) | θ value | 3.08544e-08 (rank : 47) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N0X7, O60349, Q86Y67, Q9H1T2, Q9H1T3 | Gene names | SPG20, KIAA0610, TAHCCP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spartin (Trans-activated by hepatitis C virus core protein 1). | |||||
|
RUVB2_HUMAN
|
||||||
| NC score | 0.131961 (rank : 60) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y230, Q6PK27, Q9Y361 | Gene names | RUVBL2, TIP48, TIP49B | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (48 kDa TATA box-binding protein-interacting protein) (48 kDa TBP-interacting protein) (TIP49b) (Repressing pontin 52) (Reptin 52) (51 kDa erythrocyte cytosolic protein) (ECP-51) (TIP60-associated protein 54-beta) (TAP54-beta). | |||||
|
RUVB2_MOUSE
|
||||||
| NC score | 0.125805 (rank : 61) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WTM5 | Gene names | Ruvbl2 | |||
|
Domain Architecture |

|
|||||
| Description | RuvB-like 2 (EC 3.6.1.-) (p47 protein). | |||||
|
RFC1_MOUSE
|
||||||
| NC score | 0.121034 (rank : 62) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
RFC1_HUMAN
|
||||||
| NC score | 0.111310 (rank : 63) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
KAD6_HUMAN
|
||||||
| NC score | 0.109915 (rank : 64) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y3D8 | Gene names | AK6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
THNSL_MOUSE
|
||||||
| NC score | 0.104160 (rank : 65) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BH55 | Gene names | Thnsl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
THNSL_HUMAN
|
||||||
| NC score | 0.097123 (rank : 66) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IYQ7, Q5VV21 | Gene names | THNSL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonine synthase-like 1. | |||||
|
RFC4_HUMAN
|
||||||
| NC score | 0.092456 (rank : 67) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35249, Q6FHX7 | Gene names | RFC4 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 4 (Replication factor C 37 kDa subunit) (RF-C 37 kDa subunit) (RFC37) (Activator 1 37 kDa subunit) (A1 37 kDa subunit). | |||||
|
KAD6_MOUSE
|
||||||
| NC score | 0.087678 (rank : 68) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VCP8 | Gene names | Ak6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate kinase isoenzyme 6 (EC 2.7.4.3) (ATP-AMP transphosphorylase 6). | |||||
|
RFC4_MOUSE
|
||||||
| NC score | 0.082292 (rank : 69) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99J62 | Gene names | Rfc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Replication factor C subunit 4. | |||||
|
RFC5_HUMAN
|
||||||
| NC score | 0.054754 (rank : 70) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P40937 | Gene names | RFC5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
RFC5_MOUSE
|
||||||
| NC score | 0.054032 (rank : 71) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9D0F6 | Gene names | Rfc5 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 5 (Replication factor C 36 kDa subunit) (RF-C 36 kDa subunit) (RFC36) (Activator 1 36 kDa subunit) (A1 36 kDa subunit). | |||||
|
SPN1_MOUSE
|
||||||
| NC score | 0.038567 (rank : 72) | θ value | 0.21417 (rank : 59) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80W37 | Gene names | Snupn, Rnut1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Snurportin-1 (RNA U transporter 1). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.034926 (rank : 73) | θ value | 0.163984 (rank : 58) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
NIP30_MOUSE
|
||||||
| NC score | 0.026730 (rank : 74) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WE2, Q8R0C9 | Gene names | Nip30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEFA-interacting nuclear protein NIP30. | |||||
|
RAD52_MOUSE
|
||||||
| NC score | 0.024463 (rank : 75) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43352 | Gene names | Rad52 | |||
|
Domain Architecture |

|
|||||
| Description | DNA repair protein RAD52 homolog. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.021710 (rank : 76) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.020865 (rank : 77) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.020213 (rank : 78) | θ value | 0.365318 (rank : 61) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.018906 (rank : 79) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
BAT3_MOUSE
|
||||||
| NC score | 0.018302 (rank : 80) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
PFD3_MOUSE
|
||||||
| NC score | 0.017777 (rank : 81) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P61759, O55228, Q15765, Q3U6M9, Q9CPZ0 | Gene names | Vbp1, Pfdn3 | |||
|
Domain Architecture |

|
|||||
| Description | Prefoldin subunit 3 (Von Hippel-Lindau-binding protein 1) (VHL-binding protein 1) (VBP-1). | |||||
|
PFD3_HUMAN
|
||||||
| NC score | 0.017768 (rank : 82) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P61758, O55228, Q15765, Q5JT81, Q86X96 | Gene names | VBP1, PFDN3 | |||
|
Domain Architecture |

|
|||||
| Description | Prefoldin subunit 3 (Von Hippel-Lindau-binding protein 1) (VHL-binding protein 1) (VBP-1) (HIBBJ46). | |||||
|
ST5_MOUSE
|
||||||
| NC score | 0.014855 (rank : 83) | θ value | 0.62314 (rank : 64) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q924W7, Q78H54, Q8K2P3, Q924W8 | Gene names | St5, Dennd2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (DENN domain-containing protein 2B). | |||||
|
ST5_HUMAN
|
||||||
| NC score | 0.014568 (rank : 84) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78524, P78523, Q16492, Q7KYY2, Q7KZ12, Q8NE12, Q9BQQ6 | Gene names | ST5, DENND2B, HTS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (HeLa tumor suppression 1) (DENN domain-containing protein 2B). | |||||
|
LETM1_MOUSE
|
||||||
| NC score | 0.014122 (rank : 85) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z2I0, Q5PQC5, Q7TMK8, Q80ZX6, Q8CGJ3, Q8K5E5 | Gene names | Letm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper-EF-hand-containing transmembrane protein 1, mitochondrial precursor. | |||||
|
YBOX2_HUMAN
|
||||||
| NC score | 0.013052 (rank : 86) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2T7, Q8N4P0 | Gene names | YBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Y-box-binding protein 2 (Germ cell-specific Y-box-binding protein) (Contrin) (MSY2 homolog). | |||||
|
NSMA2_HUMAN
|
||||||
| NC score | 0.012845 (rank : 87) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NY59 | Gene names | SMPD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sphingomyelin phosphodiesterase 3 (EC 3.1.4.12) (Neutral sphingomyelinase 2) (Neutral sphingomyelinase II) (nSMase2) (nSMase- 2). | |||||
|
FHOD1_MOUSE
|
||||||
| NC score | 0.012340 (rank : 88) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P9Q4, Q8BMK2 | Gene names | Fhod1, Fhos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.012055 (rank : 89) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
ABCF1_HUMAN
|
||||||
| NC score | 0.011825 (rank : 90) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NE71, O14897, Q69YP6 | Gene names | ABCF1, ABC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1 (ATP-binding cassette 50) (TNF-alpha-stimulated ABC protein). | |||||
|
MCM5_MOUSE
|
||||||
| NC score | 0.011812 (rank : 91) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49718 | Gene names | Mcm5, Cdc46, Mcmd5 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM5 (CDC46 homolog) (P1-CDC46). | |||||
|
ABCF1_MOUSE
|
||||||
| NC score | 0.011737 (rank : 92) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P542, Q6NV71 | Gene names | Abcf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1. | |||||
|
RD23B_MOUSE
|
||||||
| NC score | 0.011705 (rank : 93) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.011162 (rank : 94) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
MCM5_HUMAN
|
||||||
| NC score | 0.011051 (rank : 95) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P33992, O60785, Q14578, Q9BTJ4, Q9BWL8 | Gene names | MCM5, CDC46 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM5 (CDC46 homolog) (P1-CDC46). | |||||
|
LSD1_MOUSE
|
||||||
| NC score | 0.011007 (rank : 96) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6ZQ88, Q6PB53, Q8VEA1 | Gene names | Aof2, Kiaa0601, Lsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.010878 (rank : 97) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
CND1_HUMAN
|
||||||
| NC score | 0.010499 (rank : 98) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15021, Q8N6U3 | Gene names | NCAPD2, CAPD2, CNAP1, KIAA0159 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 1 (Non-SMC condensin I complex subunit D2) (Chromosome condensation-related SMC-associated protein 1) (Chromosome-associated protein D2) (hCAP-D2) (XCAP-D2 homolog). | |||||
|
YBOX2_MOUSE
|
||||||
| NC score | 0.010455 (rank : 99) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z2C8, Q5NCW8, Q5NCW9, Q9Z2C7 | Gene names | Ybx2, Msy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Y-box-binding protein 2 (Germ cell-specific Y-box-binding protein) (FRGY2 homolog). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.010411 (rank : 100) | θ value | 0.62314 (rank : 63) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MKL2_HUMAN
|
||||||
| NC score | 0.010370 (rank : 101) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ULH7, Q68CT1, Q6UB16, Q86WW2, Q8N226 | Gene names | MKL2, KIAA1243, MRTFB | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B) (Megakaryoblastic leukemia 2). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.009996 (rank : 102) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
LBXCO_HUMAN
|
||||||
| NC score | 0.009517 (rank : 103) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P84550 | Gene names | LBXCOR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladybird homeobox corepressor 1. | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.009055 (rank : 104) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
DYHC_MOUSE
|
||||||
| NC score | 0.009036 (rank : 105) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JHU4 | Gene names | Dync1h1, Dnch1, Dnchc1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
DYHC_HUMAN
|
||||||
| NC score | 0.008982 (rank : 106) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 733 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14204, Q6DKQ7, Q8WU28, Q92814, Q9Y4G5 | Gene names | DYNC1H1, DNCH1, DNECL, KIAA0325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.007959 (rank : 107) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
OLIG1_HUMAN
|
||||||
| NC score | 0.007924 (rank : 108) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TAK6, Q7RTS0 | Gene names | OLIG1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 1 (Oligo1). | |||||
|
EAN57_HUMAN
|
||||||
| NC score | 0.007762 (rank : 109) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43247, Q6ICF2, Q9Y4V8 | Gene names | EAN57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein EAN57. | |||||
|
GSHB_MOUSE
|
||||||
| NC score | 0.007646 (rank : 110) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51855 | Gene names | Gss | |||
|
Domain Architecture |
|
|||||
| Description | Glutathione synthetase (EC 6.3.2.3) (Glutathione synthase) (GSH synthetase) (GSH-S). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.007566 (rank : 111) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CJ108_HUMAN
|
||||||
| NC score | 0.007093 (rank : 112) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N8Z3 | Gene names | C10orf108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative uncharacterized protein C10orf108. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.007043 (rank : 113) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
ZDHC8_HUMAN
|
||||||
| NC score | 0.006276 (rank : 114) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ULC8, Q6ICL1, Q6ZNF5, Q7Z6L9 | Gene names | ZDHHC8, KIAA1292, ZDHHCL1, ZNF378 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8) (Zinc finger protein 378). | |||||
|
CX039_HUMAN
|
||||||
| NC score | 0.006249 (rank : 115) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PEV8, Q8WVP6, Q96AV3 | Gene names | CXorf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein CXorf39. | |||||
|
KS6C1_HUMAN
|
||||||
| NC score | 0.006224 (rank : 116) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96S38, Q8TDD3, Q9NSF4, Q9UL66 | Gene names | RPS6KC1, RPK118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase delta-1 (EC 2.7.11.1) (52 kDa ribosomal protein S6 kinase) (Ribosomal S6 kinase-like protein with two PSK domains 118 kDa protein) (SPHK1-binding protein). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.006163 (rank : 117) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
TF3C2_HUMAN
|
||||||
| NC score | 0.005793 (rank : 118) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WUA4, Q16632, Q9BWI7 | Gene names | GTF3C2, KIAA0011 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 2 (Transcription factor IIIC-subunit beta) (TF3C-beta) (TFIIIC 110 kDa subunit) (TFIIIC110). | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.005566 (rank : 119) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
TACC3_HUMAN
|
||||||
| NC score | 0.005093 (rank : 120) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
THAP4_HUMAN
|
||||||
| NC score | 0.004643 (rank : 121) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WY91, Q53NU7, Q6GRN0, Q6IPJ3, Q9NW26, Q9Y325 | Gene names | THAP4 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 4. | |||||
|
IMMT_HUMAN
|
||||||
| NC score | 0.004203 (rank : 122) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 494 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16891, Q14539, Q15092, Q68D41, Q69HW5, Q6IBL0, Q7Z3X1, Q8TAJ5, Q9P0V2 | Gene names | IMMT, HMP, PIG4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial inner membrane protein (Mitofilin) (p87/89) (Proliferation-inducing gene 4 protein). | |||||
|
ZFHX2_HUMAN
|
||||||
| NC score | 0.004100 (rank : 123) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.002947 (rank : 124) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BRE1B_HUMAN
|
||||||
| NC score | 0.002835 (rank : 125) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
DIAP1_MOUSE
|
||||||
| NC score | 0.002696 (rank : 126) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
RAVR2_HUMAN
|
||||||
| NC score | 0.002500 (rank : 127) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCJ3, Q6P141, Q9NPV7 | Gene names | RAVER2, KIAA1579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 2 (Protein raver-2). | |||||
|
FOXL1_HUMAN
|
||||||
| NC score | 0.002446 (rank : 128) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12952, Q9H242 | Gene names | FOXL1, FKHL11, FREAC7 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein L1 (Forkhead-related protein FKHL11) (Forkhead- related transcription factor 7) (FREAC-7). | |||||
|
CO6A2_HUMAN
|
||||||
| NC score | 0.002078 (rank : 129) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P12110, Q13909, Q13910, Q13911, Q14049, Q16259, Q16597, Q9UML3 | Gene names | COL6A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(VI) chain precursor. | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.001848 (rank : 130) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
PDE9A_HUMAN
|
||||||
| NC score | 0.001796 (rank : 131) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O76083, O75490, O75491, O95225, Q86SF7, Q86SI6, Q86SJ3, Q86WN3, Q86WN4, Q86WN5, Q86WN6, Q86WN7, Q86WN8, Q86WN9, Q86WP0 | Gene names | PDE9A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.001782 (rank : 132) | θ value | 0.813845 (rank : 66) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.001209 (rank : 133) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.001209 (rank : 134) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
SUHW3_MOUSE
|
||||||
| NC score | -0.000601 (rank : 135) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 697 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P3Y5, Q6ZPM2, Q8K145 | Gene names | Suhw3, Kiaa1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 3. | |||||
|
EHMT2_HUMAN
|
||||||
| NC score | -0.001446 (rank : 136) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
TTK_MOUSE
|
||||||
| NC score | -0.001729 (rank : 137) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35761 | Gene names | Ttk, Esk | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase TTK (EC 2.7.12.1) (ESK) (PYT). | |||||
|
M3K6_HUMAN
|
||||||
| NC score | -0.001959 (rank : 138) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95382 | Gene names | MAP3K6, MAPKKK6 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 6 (EC 2.7.11.25). | |||||
|
LAP4_MOUSE
|
||||||
| NC score | -0.002074 (rank : 139) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
MK15_HUMAN
|
||||||
| NC score | -0.002079 (rank : 140) | θ value | 0.365318 (rank : 60) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TD08, Q2TCF9, Q8N362 | Gene names | MAPK15, ERK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 15 (EC 2.7.11.24) (Extracellular signal-regulated kinase 8). | |||||
|
M3K15_HUMAN
|
||||||
| NC score | -0.002084 (rank : 141) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 914 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6ZN16, Q5JPR4, Q6ZMV3 | Gene names | MAP3K15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 15 (EC 2.7.11.25) (MAPK/ERK kinase kinase 15) (MEK kinase 15) (MEKK 15). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | -0.002236 (rank : 142) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
TTK_HUMAN
|
||||||
| NC score | -0.002525 (rank : 143) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P33981 | Gene names | TTK, MPS1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase TTK (EC 2.7.12.1) (Phosphotyrosine picked threonine-protein kinase) (PYT). | |||||
|
KLF12_MOUSE
|
||||||
| NC score | -0.005302 (rank : 144) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 138 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35738 | Gene names | Klf12, Ap2rep | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 12 (Transcriptional repressor AP-2rep). | |||||