Please be patient as the page loads
|
CND1_HUMAN
|
||||||
| SwissProt Accessions | Q15021, Q8N6U3 | Gene names | NCAPD2, CAPD2, CNAP1, KIAA0159 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 1 (Non-SMC condensin I complex subunit D2) (Chromosome condensation-related SMC-associated protein 1) (Chromosome-associated protein D2) (hCAP-D2) (XCAP-D2 homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CND1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15021, Q8N6U3 | Gene names | NCAPD2, CAPD2, CNAP1, KIAA0159 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 1 (Non-SMC condensin I complex subunit D2) (Chromosome condensation-related SMC-associated protein 1) (Chromosome-associated protein D2) (hCAP-D2) (XCAP-D2 homolog). | |||||
|
CND1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.990366 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K2Z4, Q8R0F2, Q8R3H0, Q922B7, Q923A3, Q9CS25, Q9CT32 | Gene names | Ncapd2, Capd2, Cnap1 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 1 (Non-SMC condensin I complex subunit D2) (Chromosome condensation-related SMC-associated protein 1) (Chromosome-associated protein D2) (mCAP-D2) (XCAP-D2 homolog). | |||||
|
K0056_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 3) | NC score | 0.319483 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZQK0, Q9CS21 | Gene names | Kiaa0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
K0056_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 4) | NC score | 0.316581 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42695 | Gene names | KIAA0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
KU70_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 5) | NC score | 0.081579 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P12956, Q9UCQ2, Q9UCQ3 | Gene names | XRCC6, G22P1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase 2 subunit 1 (EC 3.6.1.-) (ATP-dependent DNA helicase II 70 kDa subunit) (Lupus Ku autoantigen protein p70) (Ku70) (70 kDa subunit of Ku antigen) (Thyroid-lupus autoantigen) (TLAA) (CTC box-binding factor 75 kDa subunit) (CTCBF) (CTC75) (DNA-repair protein XRCC6). | |||||
|
KU70_MOUSE
|
||||||
| θ value | 0.813845 (rank : 6) | NC score | 0.068901 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23475, O88212, Q62027, Q62382, Q62453 | Gene names | Xrcc6, G22p1, Ku70 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase 2 subunit 1 (EC 3.6.1.-) (ATP-dependent DNA helicase II 70 kDa subunit) (Ku autoantigen protein p70 homolog) (Ku70) (CTC box-binding factor 75 kDa subunit) (CTCBF) (CTC75) (DNA- repair protein XRCC6). | |||||
|
CR054_HUMAN
|
||||||
| θ value | 1.06291 (rank : 7) | NC score | 0.046637 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IYD9, Q6MZU3, Q6ZTL6 | Gene names | C18orf54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf54 precursor. | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 8) | NC score | 0.005310 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 1.38821 (rank : 9) | NC score | 0.005502 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
B4GN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 10) | NC score | 0.041481 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00973 | Gene names | B4GALNT1, GALGT, SIAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4 N-acetylgalactosaminyltransferase 1 (EC 2.4.1.92) ((N- acetylneuraminyl)-galactosylglucosylceramide) (GM2/GD2 synthase) (GalNAc-T). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 11) | NC score | 0.007171 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
RASF5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 12) | NC score | 0.032129 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WWW0, Q5SY32, Q8WWV9, Q8WXF4, Q9BT99 | Gene names | RASSF5, NORE1, RAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras association domain-containing family protein 5 (New ras effector 1) (Regulator for cell adhesion and polarization enriched in lymphoid tissues) (RAPL). | |||||
|
TRI10_MOUSE
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.014486 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WUH5, Q80WA9, Q9CY03 | Gene names | Trim10, Herf1, Rnf9 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 10 (RING finger protein 9) (Hematopoietic RING finger 1). | |||||
|
RASF5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.030616 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5EBH1, O70407, Q6KAR0, Q8C2E8 | Gene names | Rassf5, Nore1, Rapl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras association domain-containing family protein 5 (New ras effector 1) (Regulator for cell adhesion and polarization enriched in lymphoid tissues) (RAPL). | |||||
|
TRI10_HUMAN
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.014073 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UDY6, Q86Z08, Q96QB6, Q9C023, Q9C024 | Gene names | TRIM10, RFB30, RNF9 | |||
|
Domain Architecture |
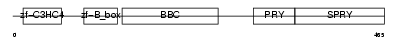
|
|||||
| Description | Tripartite motif-containing protein 10 (RING finger protein 9) (B30- RING finger protein). | |||||
|
CN021_MOUSE
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.026860 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BMC4, Q3TW87, Q8BKR9, Q8BYV4, Q8VEF9, Q9D0C6, Q9D5A8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf21 homolog. | |||||
|
COPB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.031115 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53618, Q6GTT7, Q9NTK2, Q9UNW7 | Gene names | COPB1, COPB | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
COPB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.031097 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JIF7 | Gene names | Copb1, Copb | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
SPAG5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.013988 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96R06, O95213, Q9BWE8, Q9NT17, Q9UFE6 | Gene names | SPAG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Astrin) (Mitotic spindle-associated protein p126) (MAP126) (Deepest). | |||||
|
SPAST_MOUSE
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.010499 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
MEFV_MOUSE
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.019401 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JJ26 | Gene names | Mefv | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
PDXL2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.021896 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
UBP4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.012709 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35123, O54704 | Gene names | Usp4, Unp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein). | |||||
|
ARID2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.018246 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
NEK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.002671 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
PDE2A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.008632 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00408 | Gene names | PDE2A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
TAB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.015414 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
TRI66_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.009983 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
VANG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.015281 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TAA9, Q86WG8, Q8N559 | Gene names | VANGL1, STB2 | |||
|
Domain Architecture |

|
|||||
| Description | Vang-like protein 1 (Van Gogh-like protein 1) (Strabismus 2) (Loop- tail protein 2 homolog) (LPP2). | |||||
|
CR025_MOUSE
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.020481 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BH50, Q3TQ72, Q8BU43 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf25 homolog. | |||||
|
DHRS3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.019129 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75911, Q6UY38, Q9BUC8 | Gene names | DHRS3 | |||
|
Domain Architecture |
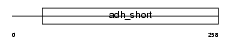
|
|||||
| Description | Short-chain dehydrogenase/reductase 3 (EC 1.1.-.-) (Retinal short- chain dehydrogenase/reductase 1) (retSDR1) (DD83.1). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.003784 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
NIBA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.016086 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZQ8, Q5TEM8, Q8TEI5, Q9H593, Q9H9Y8, Q9HCB9 | Gene names | C1orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban protein. | |||||
|
PKD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.009251 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
TENA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.002939 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24821, Q14583, Q15567 | Gene names | TNC, HXB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Myotendinous antigen) (Glioma- associated-extracellular matrix antigen) (GP 150-225). | |||||
|
LBN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.011339 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K1G2, Q8BRF3 | Gene names | Evc2, Lbn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Limbin. | |||||
|
UBP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.010438 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
CND1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15021, Q8N6U3 | Gene names | NCAPD2, CAPD2, CNAP1, KIAA0159 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 1 (Non-SMC condensin I complex subunit D2) (Chromosome condensation-related SMC-associated protein 1) (Chromosome-associated protein D2) (hCAP-D2) (XCAP-D2 homolog). | |||||
|
CND1_MOUSE
|
||||||
| NC score | 0.990366 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K2Z4, Q8R0F2, Q8R3H0, Q922B7, Q923A3, Q9CS25, Q9CT32 | Gene names | Ncapd2, Capd2, Cnap1 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 1 (Non-SMC condensin I complex subunit D2) (Chromosome condensation-related SMC-associated protein 1) (Chromosome-associated protein D2) (mCAP-D2) (XCAP-D2 homolog). | |||||
|
K0056_MOUSE
|
||||||
| NC score | 0.319483 (rank : 3) | θ value | 8.11959e-09 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZQK0, Q9CS21 | Gene names | Kiaa0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
K0056_HUMAN
|
||||||
| NC score | 0.316581 (rank : 4) | θ value | 2.36244e-08 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42695 | Gene names | KIAA0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
KU70_HUMAN
|
||||||
| NC score | 0.081579 (rank : 5) | θ value | 0.0961366 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P12956, Q9UCQ2, Q9UCQ3 | Gene names | XRCC6, G22P1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase 2 subunit 1 (EC 3.6.1.-) (ATP-dependent DNA helicase II 70 kDa subunit) (Lupus Ku autoantigen protein p70) (Ku70) (70 kDa subunit of Ku antigen) (Thyroid-lupus autoantigen) (TLAA) (CTC box-binding factor 75 kDa subunit) (CTCBF) (CTC75) (DNA-repair protein XRCC6). | |||||
|
KU70_MOUSE
|
||||||
| NC score | 0.068901 (rank : 6) | θ value | 0.813845 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23475, O88212, Q62027, Q62382, Q62453 | Gene names | Xrcc6, G22p1, Ku70 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase 2 subunit 1 (EC 3.6.1.-) (ATP-dependent DNA helicase II 70 kDa subunit) (Ku autoantigen protein p70 homolog) (Ku70) (CTC box-binding factor 75 kDa subunit) (CTCBF) (CTC75) (DNA- repair protein XRCC6). | |||||
|
CR054_HUMAN
|
||||||
| NC score | 0.046637 (rank : 7) | θ value | 1.06291 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IYD9, Q6MZU3, Q6ZTL6 | Gene names | C18orf54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf54 precursor. | |||||
|
B4GN1_HUMAN
|
||||||
| NC score | 0.041481 (rank : 8) | θ value | 1.81305 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00973 | Gene names | B4GALNT1, GALGT, SIAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4 N-acetylgalactosaminyltransferase 1 (EC 2.4.1.92) ((N- acetylneuraminyl)-galactosylglucosylceramide) (GM2/GD2 synthase) (GalNAc-T). | |||||
|
RASF5_HUMAN
|
||||||
| NC score | 0.032129 (rank : 9) | θ value | 1.81305 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WWW0, Q5SY32, Q8WWV9, Q8WXF4, Q9BT99 | Gene names | RASSF5, NORE1, RAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras association domain-containing family protein 5 (New ras effector 1) (Regulator for cell adhesion and polarization enriched in lymphoid tissues) (RAPL). | |||||
|
COPB_HUMAN
|
||||||
| NC score | 0.031115 (rank : 10) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53618, Q6GTT7, Q9NTK2, Q9UNW7 | Gene names | COPB1, COPB | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
COPB_MOUSE
|
||||||
| NC score | 0.031097 (rank : 11) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JIF7 | Gene names | Copb1, Copb | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
RASF5_MOUSE
|
||||||
| NC score | 0.030616 (rank : 12) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5EBH1, O70407, Q6KAR0, Q8C2E8 | Gene names | Rassf5, Nore1, Rapl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras association domain-containing family protein 5 (New ras effector 1) (Regulator for cell adhesion and polarization enriched in lymphoid tissues) (RAPL). | |||||
|
CN021_MOUSE
|
||||||
| NC score | 0.026860 (rank : 13) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BMC4, Q3TW87, Q8BKR9, Q8BYV4, Q8VEF9, Q9D0C6, Q9D5A8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf21 homolog. | |||||
|
PDXL2_MOUSE
|
||||||
| NC score | 0.021896 (rank : 14) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
CR025_MOUSE
|
||||||
| NC score | 0.020481 (rank : 15) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BH50, Q3TQ72, Q8BU43 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf25 homolog. | |||||
|
MEFV_MOUSE
|
||||||
| NC score | 0.019401 (rank : 16) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JJ26 | Gene names | Mefv | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
DHRS3_HUMAN
|
||||||
| NC score | 0.019129 (rank : 17) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75911, Q6UY38, Q9BUC8 | Gene names | DHRS3 | |||
|
Domain Architecture |

|
|||||
| Description | Short-chain dehydrogenase/reductase 3 (EC 1.1.-.-) (Retinal short- chain dehydrogenase/reductase 1) (retSDR1) (DD83.1). | |||||
|
ARID2_HUMAN
|
||||||
| NC score | 0.018246 (rank : 18) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
NIBA_HUMAN
|
||||||
| NC score | 0.016086 (rank : 19) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZQ8, Q5TEM8, Q8TEI5, Q9H593, Q9H9Y8, Q9HCB9 | Gene names | C1orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban protein. | |||||
|
TAB2_MOUSE
|
||||||
| NC score | 0.015414 (rank : 20) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
VANG1_HUMAN
|
||||||
| NC score | 0.015281 (rank : 21) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TAA9, Q86WG8, Q8N559 | Gene names | VANGL1, STB2 | |||
|
Domain Architecture |

|
|||||
| Description | Vang-like protein 1 (Van Gogh-like protein 1) (Strabismus 2) (Loop- tail protein 2 homolog) (LPP2). | |||||
|
TRI10_MOUSE
|
||||||
| NC score | 0.014486 (rank : 22) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WUH5, Q80WA9, Q9CY03 | Gene names | Trim10, Herf1, Rnf9 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 10 (RING finger protein 9) (Hematopoietic RING finger 1). | |||||
|
TRI10_HUMAN
|
||||||
| NC score | 0.014073 (rank : 23) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UDY6, Q86Z08, Q96QB6, Q9C023, Q9C024 | Gene names | TRIM10, RFB30, RNF9 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 10 (RING finger protein 9) (B30- RING finger protein). | |||||
|
SPAG5_HUMAN
|
||||||
| NC score | 0.013988 (rank : 24) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96R06, O95213, Q9BWE8, Q9NT17, Q9UFE6 | Gene names | SPAG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Astrin) (Mitotic spindle-associated protein p126) (MAP126) (Deepest). | |||||
|
UBP4_MOUSE
|
||||||
| NC score | 0.012709 (rank : 25) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35123, O54704 | Gene names | Usp4, Unp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein). | |||||
|
LBN_MOUSE
|
||||||
| NC score | 0.011339 (rank : 26) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K1G2, Q8BRF3 | Gene names | Evc2, Lbn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Limbin. | |||||
|
SPAST_MOUSE
|
||||||
| NC score | 0.010499 (rank : 27) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
UBP4_HUMAN
|
||||||
| NC score | 0.010438 (rank : 28) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.009983 (rank : 29) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
PKD1_HUMAN
|
||||||
| NC score | 0.009251 (rank : 30) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
PDE2A_HUMAN
|
||||||
| NC score | 0.008632 (rank : 31) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00408 | Gene names | PDE2A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.007171 (rank : 32) | θ value | 1.81305 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.005502 (rank : 33) | θ value | 1.38821 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.005310 (rank : 34) | θ value | 1.06291 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.003784 (rank : 35) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
TENA_HUMAN
|
||||||
| NC score | 0.002939 (rank : 36) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24821, Q14583, Q15567 | Gene names | TNC, HXB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Myotendinous antigen) (Glioma- associated-extracellular matrix antigen) (GP 150-225). | |||||
|
NEK1_MOUSE
|
||||||
| NC score | 0.002671 (rank : 37) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||