Please be patient as the page loads
|
NSMA2_HUMAN
|
||||||
| SwissProt Accessions | Q9NY59 | Gene names | SMPD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sphingomyelin phosphodiesterase 3 (EC 3.1.4.12) (Neutral sphingomyelinase 2) (Neutral sphingomyelinase II) (nSMase2) (nSMase- 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NSMA2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NY59 | Gene names | SMPD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sphingomyelin phosphodiesterase 3 (EC 3.1.4.12) (Neutral sphingomyelinase 2) (Neutral sphingomyelinase II) (nSMase2) (nSMase- 2). | |||||
|
NSMA2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.927819 (rank : 2) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JJY3, Q3UTQ5 | Gene names | smpd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sphingomyelin phosphodiesterase 3 (EC 3.1.4.12) (Neutral sphingomyelinase 2) (Neutral sphingomyelinase II) (nSMase2) (nSMase- 2). | |||||
|
BEGIN_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 3) | NC score | 0.074308 (rank : 3) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BUH8, Q9NPU3, Q9P282 | Gene names | BEGAIN, KIAA1446 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-enriched guanylate kinase-associated protein. | |||||
|
CO4A4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 4) | NC score | 0.045010 (rank : 7) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P53420 | Gene names | COL4A4 | |||
|
Domain Architecture |
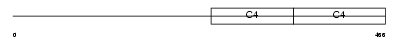
|
|||||
| Description | Collagen alpha-4(IV) chain precursor. | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 5) | NC score | 0.046614 (rank : 6) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 6) | NC score | 0.020433 (rank : 18) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ADIPO_HUMAN
|
||||||
| θ value | 1.38821 (rank : 7) | NC score | 0.029466 (rank : 11) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15848 | Gene names | ADIPOQ, ACDC, ACRP30, APM1, GBP28 | |||
|
Domain Architecture |
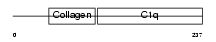
|
|||||
| Description | Adiponectin precursor (Adipocyte, C1q and collagen domain-containing protein) (30 kDa adipocyte complement-related protein) (Adipocyte complement-related 30 kDa protein) (ACRP30) (Adipose most abundant gene transcript 1) (apM-1) (Gelatin-binding protein). | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 8) | NC score | 0.031674 (rank : 10) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
BAI1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 9) | NC score | 0.011560 (rank : 22) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UHD1, Q3UH36, Q8CGM0 | Gene names | Bai1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
CJ107_MOUSE
|
||||||
| θ value | 1.81305 (rank : 10) | NC score | 0.069364 (rank : 4) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CDT7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf107 homolog. | |||||
|
CO9A1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 11) | NC score | 0.025592 (rank : 15) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
SPAST_MOUSE
|
||||||
| θ value | 2.36792 (rank : 12) | NC score | 0.012845 (rank : 20) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
ABL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.005091 (rank : 27) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
ADIPO_MOUSE
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.026363 (rank : 14) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60994, Q62400, Q9DC68 | Gene names | Adipoq, Acdc, Acrp30, Apm1 | |||
|
Domain Architecture |
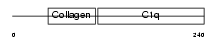
|
|||||
| Description | Adiponectin precursor (Adipocyte, C1q and collagen domain-containing protein) (30 kDa adipocyte complement-related protein) (Adipocyte complement-related 30 kDa protein) (ACRP30) (Adipocyte-specific protein AdipoQ). | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.028029 (rank : 13) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
CO4A2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.029219 (rank : 12) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
MARCO_MOUSE
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.040648 (rank : 9) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60754 | Gene names | Marco | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure). | |||||
|
CO5A3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.042865 (rank : 8) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P25940, Q9NZQ6 | Gene names | COL5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(V) chain precursor. | |||||
|
NCOA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.010228 (rank : 23) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 20) | NC score | 0.015165 (rank : 19) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SERA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.022278 (rank : 16) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43175, Q5SZU3, Q9BQ01 | Gene names | PHGDH, PGDH3 | |||
|
Domain Architecture |
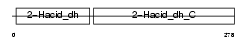
|
|||||
| Description | D-3-phosphoglycerate dehydrogenase (EC 1.1.1.95) (3-PGDH). | |||||
|
SERA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.022272 (rank : 17) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61753, Q75SV9 | Gene names | Phgdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | D-3-phosphoglycerate dehydrogenase (EC 1.1.1.95) (3-PGDH) (A10). | |||||
|
ZN688_HUMAN
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.000527 (rank : 28) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WV14, O75701, Q8IW91, Q96MN0 | Gene names | ZNF688 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 688. | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.008360 (rank : 25) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
JADE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.007140 (rank : 26) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6IE81, Q4W5D5, Q6ZSL7, Q8NC41, Q96JL8, Q96SQ1, Q9H692 | Gene names | PHF17, JADE1, KIAA1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.008951 (rank : 24) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
RRMJ3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.012843 (rank : 21) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DBE9, Q3ULI1, Q921I7 | Gene names | Ftsj3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2'-O-)- methyltransferase 3). | |||||
|
NEUM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.063307 (rank : 5) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
NSMA2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NY59 | Gene names | SMPD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sphingomyelin phosphodiesterase 3 (EC 3.1.4.12) (Neutral sphingomyelinase 2) (Neutral sphingomyelinase II) (nSMase2) (nSMase- 2). | |||||
|
NSMA2_MOUSE
|
||||||
| NC score | 0.927819 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JJY3, Q3UTQ5 | Gene names | smpd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sphingomyelin phosphodiesterase 3 (EC 3.1.4.12) (Neutral sphingomyelinase 2) (Neutral sphingomyelinase II) (nSMase2) (nSMase- 2). | |||||
|
BEGIN_HUMAN
|
||||||
| NC score | 0.074308 (rank : 3) | θ value | 0.0431538 (rank : 3) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BUH8, Q9NPU3, Q9P282 | Gene names | BEGAIN, KIAA1446 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-enriched guanylate kinase-associated protein. | |||||
|
CJ107_MOUSE
|
||||||
| NC score | 0.069364 (rank : 4) | θ value | 1.81305 (rank : 10) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CDT7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf107 homolog. | |||||
|
NEUM_HUMAN
|
||||||
| NC score | 0.063307 (rank : 5) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.046614 (rank : 6) | θ value | 0.813845 (rank : 5) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
CO4A4_HUMAN
|
||||||
| NC score | 0.045010 (rank : 7) | θ value | 0.125558 (rank : 4) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P53420 | Gene names | COL4A4 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-4(IV) chain precursor. | |||||
|
CO5A3_HUMAN
|
||||||
| NC score | 0.042865 (rank : 8) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P25940, Q9NZQ6 | Gene names | COL5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(V) chain precursor. | |||||
|
MARCO_MOUSE
|
||||||
| NC score | 0.040648 (rank : 9) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60754 | Gene names | Marco | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure). | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.031674 (rank : 10) | θ value | 1.38821 (rank : 8) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
ADIPO_HUMAN
|
||||||
| NC score | 0.029466 (rank : 11) | θ value | 1.38821 (rank : 7) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15848 | Gene names | ADIPOQ, ACDC, ACRP30, APM1, GBP28 | |||
|
Domain Architecture |

|
|||||
| Description | Adiponectin precursor (Adipocyte, C1q and collagen domain-containing protein) (30 kDa adipocyte complement-related protein) (Adipocyte complement-related 30 kDa protein) (ACRP30) (Adipose most abundant gene transcript 1) (apM-1) (Gelatin-binding protein). | |||||
|
CO4A2_HUMAN
|
||||||
| NC score | 0.029219 (rank : 12) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.028029 (rank : 13) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
ADIPO_MOUSE
|
||||||
| NC score | 0.026363 (rank : 14) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60994, Q62400, Q9DC68 | Gene names | Adipoq, Acdc, Acrp30, Apm1 | |||
|
Domain Architecture |

|
|||||
| Description | Adiponectin precursor (Adipocyte, C1q and collagen domain-containing protein) (30 kDa adipocyte complement-related protein) (Adipocyte complement-related 30 kDa protein) (ACRP30) (Adipocyte-specific protein AdipoQ). | |||||
|
CO9A1_MOUSE
|
||||||
| NC score | 0.025592 (rank : 15) | θ value | 1.81305 (rank : 11) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
SERA_HUMAN
|
||||||
| NC score | 0.022278 (rank : 16) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43175, Q5SZU3, Q9BQ01 | Gene names | PHGDH, PGDH3 | |||
|
Domain Architecture |

|
|||||
| Description | D-3-phosphoglycerate dehydrogenase (EC 1.1.1.95) (3-PGDH). | |||||
|
SERA_MOUSE
|
||||||
| NC score | 0.022272 (rank : 17) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61753, Q75SV9 | Gene names | Phgdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | D-3-phosphoglycerate dehydrogenase (EC 1.1.1.95) (3-PGDH) (A10). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.020433 (rank : 18) | θ value | 0.813845 (rank : 6) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.015165 (rank : 19) | θ value | 6.88961 (rank : 20) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SPAST_MOUSE
|
||||||
| NC score | 0.012845 (rank : 20) | θ value | 2.36792 (rank : 12) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
RRMJ3_MOUSE
|
||||||
| NC score | 0.012843 (rank : 21) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DBE9, Q3ULI1, Q921I7 | Gene names | Ftsj3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2'-O-)- methyltransferase 3). | |||||
|
BAI1_MOUSE
|
||||||
| NC score | 0.011560 (rank : 22) | θ value | 1.81305 (rank : 9) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UHD1, Q3UH36, Q8CGM0 | Gene names | Bai1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
NCOA1_MOUSE
|
||||||
| NC score | 0.010228 (rank : 23) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.008951 (rank : 24) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.008360 (rank : 25) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
JADE1_HUMAN
|
||||||
| NC score | 0.007140 (rank : 26) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6IE81, Q4W5D5, Q6ZSL7, Q8NC41, Q96JL8, Q96SQ1, Q9H692 | Gene names | PHF17, JADE1, KIAA1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
ABL1_HUMAN
|
||||||
| NC score | 0.005091 (rank : 27) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
ZN688_HUMAN
|
||||||
| NC score | 0.000527 (rank : 28) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WV14, O75701, Q8IW91, Q96MN0 | Gene names | ZNF688 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 688. | |||||