Please be patient as the page loads
|
PDE9A_HUMAN
|
||||||
| SwissProt Accessions | O76083, O75490, O75491, O95225, Q86SF7, Q86SI6, Q86SJ3, Q86WN3, Q86WN4, Q86WN5, Q86WN6, Q86WN7, Q86WN8, Q86WN9, Q86WP0 | Gene names | PDE9A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PDE9A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | O76083, O75490, O75491, O95225, Q86SF7, Q86SI6, Q86SJ3, Q86WN3, Q86WN4, Q86WN5, Q86WN6, Q86WN7, Q86WN8, Q86WN9, Q86WP0 | Gene names | PDE9A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE9A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.997194 (rank : 2) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O70628 | Gene names | Pde9a, Pde8b | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE4D_MOUSE
|
||||||
| θ value | 2.04573e-44 (rank : 3) | NC score | 0.919695 (rank : 10) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q01063, Q6TRH9, Q8C4Q7, Q8CG05 | Gene names | Pde4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3). | |||||
|
PDE4B_HUMAN
|
||||||
| θ value | 2.67181e-44 (rank : 4) | NC score | 0.920494 (rank : 9) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q07343, O15443 | Gene names | PDE4B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4B (EC 3.1.4.17) (DPDE4) (PDE32). | |||||
|
PDE4D_HUMAN
|
||||||
| θ value | 2.67181e-44 (rank : 5) | NC score | 0.919051 (rank : 13) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q08499, O43433, Q13549, Q13550, Q13551, Q7Z2L8, Q8IV84, Q8IVA9, Q8IVD2, Q8IVD3, Q9HCX7 | Gene names | PDE4D | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3) (PDE43). | |||||
|
PDE4A_HUMAN
|
||||||
| θ value | 3.48949e-44 (rank : 6) | NC score | 0.919377 (rank : 12) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27815, O75522, O76092, Q16255, Q16691, Q8WUQ3 | Gene names | PDE4A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17) (DPDE2) (PDE46). | |||||
|
PDE4A_MOUSE
|
||||||
| θ value | 3.48949e-44 (rank : 7) | NC score | 0.919635 (rank : 11) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O89084, Q8R078, Q9JHQ4, Q9QX48, Q9QX49, Q9QXI8 | Gene names | Pde4a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17). | |||||
|
PDE8A_HUMAN
|
||||||
| θ value | 1.46607e-42 (rank : 8) | NC score | 0.916591 (rank : 15) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O60658, Q969I1, Q96PC9, Q96PD0, Q96PD1, Q9UMB7 | Gene names | PDE8A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17). | |||||
|
PDE4C_HUMAN
|
||||||
| θ value | 2.50074e-42 (rank : 9) | NC score | 0.922384 (rank : 8) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08493, Q9UN44, Q9UN45, Q9UN46, Q9UPJ6 | Gene names | PDE4C | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17) (DPDE1) (PDE21). | |||||
|
PDE4C_MOUSE
|
||||||
| θ value | 8.04462e-41 (rank : 10) | NC score | 0.923592 (rank : 7) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q3UEI1, Q8K0P4 | Gene names | Pde4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17). | |||||
|
PDE8B_HUMAN
|
||||||
| θ value | 2.34062e-40 (rank : 11) | NC score | 0.913086 (rank : 19) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95263, Q86XK8, Q8IUJ7, Q8IUJ8, Q8IUJ9, Q8IUK0, Q8N3T2 | Gene names | PDE8B | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8B (EC 3.1.4.17) (HSPDE8B). | |||||
|
PDE11_MOUSE
|
||||||
| θ value | 8.89434e-40 (rank : 12) | NC score | 0.889898 (rank : 26) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P0C1Q2 | Gene names | Pde11a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE8A_MOUSE
|
||||||
| θ value | 1.98146e-39 (rank : 13) | NC score | 0.912621 (rank : 20) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88502 | Gene names | Pde8a, Pde8 | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17) (MMPDE8). | |||||
|
PDE11_HUMAN
|
||||||
| θ value | 3.37985e-39 (rank : 14) | NC score | 0.889931 (rank : 25) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HCR9, Q53T16, Q96S76, Q9GZY7, Q9HB46, Q9NY45 | Gene names | PDE11A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE7A_HUMAN
|
||||||
| θ value | 1.28434e-38 (rank : 15) | NC score | 0.931451 (rank : 3) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13946, O15380 | Gene names | PDE7A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (HCP1) (TM22). | |||||
|
PDE7A_MOUSE
|
||||||
| θ value | 3.73686e-38 (rank : 16) | NC score | 0.930596 (rank : 4) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70453, Q9ERB3 | Gene names | Pde7a | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (P2A). | |||||
|
PDE2A_HUMAN
|
||||||
| θ value | 4.88048e-38 (rank : 17) | NC score | 0.900231 (rank : 24) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00408 | Gene names | PDE2A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE2A_MOUSE
|
||||||
| θ value | 4.88048e-38 (rank : 18) | NC score | 0.901064 (rank : 23) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q922S4, Q8K2U1 | Gene names | Pde2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE1B_MOUSE
|
||||||
| θ value | 2.42216e-37 (rank : 19) | NC score | 0.917216 (rank : 14) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q01065, O35384 | Gene names | Pde1b, Pde1b1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE7B_MOUSE
|
||||||
| θ value | 9.20422e-37 (rank : 20) | NC score | 0.929270 (rank : 5) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QXQ1 | Gene names | Pde7b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE1B_HUMAN
|
||||||
| θ value | 1.20211e-36 (rank : 21) | NC score | 0.914837 (rank : 17) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q01064, Q92825 | Gene names | PDE1B, PDE1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE1A_MOUSE
|
||||||
| θ value | 1.57e-36 (rank : 22) | NC score | 0.915018 (rank : 16) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61481, O35388 | Gene names | Pde1a | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE). | |||||
|
PDE7B_HUMAN
|
||||||
| θ value | 3.4976e-36 (rank : 23) | NC score | 0.928468 (rank : 6) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NP56 | Gene names | PDE7B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE1A_HUMAN
|
||||||
| θ value | 1.32909e-35 (rank : 24) | NC score | 0.913256 (rank : 18) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P54750, Q86VZ0, Q9C0K8, Q9C0K9, Q9C0L0, Q9C0L1, Q9C0L2, Q9C0L3, Q9C0L4, Q9UFX3 | Gene names | PDE1A | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE) (hCam-1). | |||||
|
PDE1C_HUMAN
|
||||||
| θ value | 6.59618e-35 (rank : 25) | NC score | 0.903547 (rank : 21) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
PDE1C_MOUSE
|
||||||
| θ value | 3.27365e-34 (rank : 26) | NC score | 0.902329 (rank : 22) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
PDE5A_MOUSE
|
||||||
| θ value | 2.34606e-32 (rank : 27) | NC score | 0.871769 (rank : 31) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CG03 | Gene names | Pde5a, Pde5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE5A_HUMAN
|
||||||
| θ value | 3.06405e-32 (rank : 28) | NC score | 0.869579 (rank : 32) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O76074, O75026, O75887, Q86UI0, Q86V66, Q9Y6Z6 | Gene names | PDE5A, PDE5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE6B_MOUSE
|
||||||
| θ value | 1.16434e-31 (rank : 29) | NC score | 0.847671 (rank : 37) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P23440 | Gene names | Pde6b, Mpb, Pdeb, rd | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PDE6A_HUMAN
|
||||||
| θ value | 1.28732e-30 (rank : 30) | NC score | 0.850441 (rank : 35) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16499 | Gene names | PDE6A, PDEA | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha) (PDE V-B1). | |||||
|
PDE6B_HUMAN
|
||||||
| θ value | 1.28732e-30 (rank : 31) | NC score | 0.848812 (rank : 36) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35913, Q9BWH5 | Gene names | PDE6B, PDEB | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase beta-subunit precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PDE6C_HUMAN
|
||||||
| θ value | 1.08979e-29 (rank : 32) | NC score | 0.853302 (rank : 34) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P51160, Q5VY29 | Gene names | PDE6C, PDEA2 | |||
|
Domain Architecture |

|
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE6C_MOUSE
|
||||||
| θ value | 2.42779e-29 (rank : 33) | NC score | 0.854186 (rank : 33) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91ZQ1, Q8R0D4 | Gene names | Pde6c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE3A_HUMAN
|
||||||
| θ value | 1.57365e-28 (rank : 34) | NC score | 0.874453 (rank : 29) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14432, O60865, Q13348 | Gene names | PDE3A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PDE6A_MOUSE
|
||||||
| θ value | 3.50572e-28 (rank : 35) | NC score | 0.846012 (rank : 38) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27664 | Gene names | Pde6a, Mpa, Pdea | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha). | |||||
|
PDE3A_MOUSE
|
||||||
| θ value | 7.80994e-28 (rank : 36) | NC score | 0.873256 (rank : 30) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Z0X4 | Gene names | Pde3a | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PDE3B_HUMAN
|
||||||
| θ value | 2.96777e-27 (rank : 37) | NC score | 0.884607 (rank : 27) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13370, O00639, Q14408 | Gene names | PDE3B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase B (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase B) (CGI-PDE B) (CGIPDE1) (CGIP1). | |||||
|
PDE10_HUMAN
|
||||||
| θ value | 6.61148e-27 (rank : 38) | NC score | 0.879796 (rank : 28) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y233, Q9Y5T1 | Gene names | PDE10A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A (EC 3.1.4.17). | |||||
|
SESN1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 39) | NC score | 0.061199 (rank : 39) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58006, Q7TNF3 | Gene names | Sesn1, Pa26, Sest1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
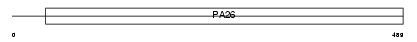
| Description | Sestrin-1 (p53-regulated protein PA26). | |||||
|
SESN1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 40) | NC score | 0.053079 (rank : 41) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6P5, Q9NV00, Q9UPD5, Q9Y6P6 | Gene names | SESN1, PA26, SEST1 | |||
|
Domain Architecture |
|
|||||
| Description | Sestrin-1 (p53-regulated protein PA26). | |||||
|
PDCD7_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 41) | NC score | 0.054919 (rank : 40) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WTY1, Q8R5D9 | Gene names | Pdcd7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death protein 7 (ES18). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 42) | NC score | 0.039144 (rank : 42) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 43) | NC score | 0.038204 (rank : 46) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 44) | NC score | 0.038709 (rank : 44) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 45) | NC score | 0.038961 (rank : 43) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
PARC_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 46) | NC score | 0.037135 (rank : 48) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 47) | NC score | 0.038377 (rank : 45) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 0.279714 (rank : 48) | NC score | 0.036697 (rank : 49) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
CING_HUMAN
|
||||||
| θ value | 0.47712 (rank : 49) | NC score | 0.028873 (rank : 62) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
RADI_HUMAN
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | 0.016916 (rank : 80) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
CCD41_MOUSE
|
||||||
| θ value | 0.62314 (rank : 51) | NC score | 0.024103 (rank : 69) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.037698 (rank : 47) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
PARC_MOUSE
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.030997 (rank : 58) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80TT8, Q8BKL9, Q8CGC0, Q8R363 | Gene names | Parc, Kiaa0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein. | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | -0.004046 (rank : 93) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.021983 (rank : 73) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CING_MOUSE
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.025808 (rank : 66) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.036120 (rank : 50) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
PDCD7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.033366 (rank : 54) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N8D1, Q96AK8, Q9Y6D7 | Gene names | PDCD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death protein 7 (ES18) (HES18). | |||||
|
TMF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.025413 (rank : 68) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
JPH3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.007104 (rank : 84) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ET77, Q9EQZ2 | Gene names | Jph3, Jp3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
LZTS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.020468 (rank : 75) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P60853 | Gene names | Lzts1, Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/Esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.022404 (rank : 72) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.019817 (rank : 76) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.025595 (rank : 67) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.031605 (rank : 57) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
PIAS1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.006752 (rank : 87) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75925, Q99751, Q9UN02 | Gene names | PIAS1, DDXBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 1 (Gu-binding protein) (GBP) (RNA helicase II-binding protein) (DEAD/H box-binding protein 1). | |||||
|
SM1L2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.022935 (rank : 71) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
SMC4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.027022 (rank : 64) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.017238 (rank : 79) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.029893 (rank : 59) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
BRE1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.026060 (rank : 65) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
BRE1B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.027058 (rank : 63) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.008927 (rank : 83) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.031939 (rank : 56) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.033227 (rank : 55) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
2A5A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.004129 (rank : 88) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PD03, Q8R1U7 | Gene names | Ppp2r5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform (PP2A, B subunit, B' alpha isoform) (PP2A, B subunit, B56 alpha isoform) (PP2A, B subunit, PR61 alpha isoform) (PP2A, B subunit, R5 alpha isoform) (PR61alpha). | |||||
|
DESM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.006961 (rank : 85) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P17661, Q15787, Q8IZR1, Q8IZR6, Q8NES2, Q8NEU6, Q8TAC4, Q8TCX2, Q8TD99, Q9UHN5, Q9UJ80 | Gene names | DES | |||
|
Domain Architecture |
|
|||||
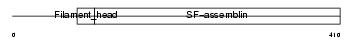
| Description | Desmin. | |||||
|
E41L2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.004000 (rank : 89) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.021145 (rank : 74) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.034163 (rank : 51) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.033789 (rank : 53) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.029271 (rank : 60) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
RADI_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.014008 (rank : 82) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
SM1L2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.023909 (rank : 70) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.006943 (rank : 86) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
SPAST_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.001796 (rank : 91) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.016849 (rank : 81) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
ANR47_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.002642 (rank : 90) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z1P7 | Gene names | Ankrd47, D17Ertd288e, Ng28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 47. | |||||
|
CD2L1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | -0.002737 (rank : 92) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
ERC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.018993 (rank : 77) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
ERC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.018894 (rank : 78) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.034101 (rank : 52) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.028956 (rank : 61) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
PDE9A_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | O76083, O75490, O75491, O95225, Q86SF7, Q86SI6, Q86SJ3, Q86WN3, Q86WN4, Q86WN5, Q86WN6, Q86WN7, Q86WN8, Q86WN9, Q86WP0 | Gene names | PDE9A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE9A_MOUSE
|
||||||
| NC score | 0.997194 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O70628 | Gene names | Pde9a, Pde8b | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE7A_HUMAN
|
||||||
| NC score | 0.931451 (rank : 3) | θ value | 1.28434e-38 (rank : 15) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13946, O15380 | Gene names | PDE7A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (HCP1) (TM22). | |||||
|
PDE7A_MOUSE
|
||||||
| NC score | 0.930596 (rank : 4) | θ value | 3.73686e-38 (rank : 16) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70453, Q9ERB3 | Gene names | Pde7a | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (P2A). | |||||
|
PDE7B_MOUSE
|
||||||
| NC score | 0.929270 (rank : 5) | θ value | 9.20422e-37 (rank : 20) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QXQ1 | Gene names | Pde7b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE7B_HUMAN
|
||||||
| NC score | 0.928468 (rank : 6) | θ value | 3.4976e-36 (rank : 23) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NP56 | Gene names | PDE7B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE4C_MOUSE
|
||||||
| NC score | 0.923592 (rank : 7) | θ value | 8.04462e-41 (rank : 10) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q3UEI1, Q8K0P4 | Gene names | Pde4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17). | |||||
|
PDE4C_HUMAN
|
||||||
| NC score | 0.922384 (rank : 8) | θ value | 2.50074e-42 (rank : 9) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08493, Q9UN44, Q9UN45, Q9UN46, Q9UPJ6 | Gene names | PDE4C | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17) (DPDE1) (PDE21). | |||||
|
PDE4B_HUMAN
|
||||||
| NC score | 0.920494 (rank : 9) | θ value | 2.67181e-44 (rank : 4) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q07343, O15443 | Gene names | PDE4B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4B (EC 3.1.4.17) (DPDE4) (PDE32). | |||||
|
PDE4D_MOUSE
|
||||||
| NC score | 0.919695 (rank : 10) | θ value | 2.04573e-44 (rank : 3) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q01063, Q6TRH9, Q8C4Q7, Q8CG05 | Gene names | Pde4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3). | |||||
|
PDE4A_MOUSE
|
||||||
| NC score | 0.919635 (rank : 11) | θ value | 3.48949e-44 (rank : 7) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O89084, Q8R078, Q9JHQ4, Q9QX48, Q9QX49, Q9QXI8 | Gene names | Pde4a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17). | |||||
|
PDE4A_HUMAN
|
||||||
| NC score | 0.919377 (rank : 12) | θ value | 3.48949e-44 (rank : 6) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27815, O75522, O76092, Q16255, Q16691, Q8WUQ3 | Gene names | PDE4A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17) (DPDE2) (PDE46). | |||||
|
PDE4D_HUMAN
|
||||||
| NC score | 0.919051 (rank : 13) | θ value | 2.67181e-44 (rank : 5) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q08499, O43433, Q13549, Q13550, Q13551, Q7Z2L8, Q8IV84, Q8IVA9, Q8IVD2, Q8IVD3, Q9HCX7 | Gene names | PDE4D | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3) (PDE43). | |||||
|
PDE1B_MOUSE
|
||||||
| NC score | 0.917216 (rank : 14) | θ value | 2.42216e-37 (rank : 19) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q01065, O35384 | Gene names | Pde1b, Pde1b1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE8A_HUMAN
|
||||||
| NC score | 0.916591 (rank : 15) | θ value | 1.46607e-42 (rank : 8) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O60658, Q969I1, Q96PC9, Q96PD0, Q96PD1, Q9UMB7 | Gene names | PDE8A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17). | |||||
|
PDE1A_MOUSE
|
||||||
| NC score | 0.915018 (rank : 16) | θ value | 1.57e-36 (rank : 22) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61481, O35388 | Gene names | Pde1a | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE). | |||||
|
PDE1B_HUMAN
|
||||||
| NC score | 0.914837 (rank : 17) | θ value | 1.20211e-36 (rank : 21) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q01064, Q92825 | Gene names | PDE1B, PDE1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE1A_HUMAN
|
||||||
| NC score | 0.913256 (rank : 18) | θ value | 1.32909e-35 (rank : 24) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P54750, Q86VZ0, Q9C0K8, Q9C0K9, Q9C0L0, Q9C0L1, Q9C0L2, Q9C0L3, Q9C0L4, Q9UFX3 | Gene names | PDE1A | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE) (hCam-1). | |||||
|
PDE8B_HUMAN
|
||||||
| NC score | 0.913086 (rank : 19) | θ value | 2.34062e-40 (rank : 11) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95263, Q86XK8, Q8IUJ7, Q8IUJ8, Q8IUJ9, Q8IUK0, Q8N3T2 | Gene names | PDE8B | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8B (EC 3.1.4.17) (HSPDE8B). | |||||
|
PDE8A_MOUSE
|
||||||
| NC score | 0.912621 (rank : 20) | θ value | 1.98146e-39 (rank : 13) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88502 | Gene names | Pde8a, Pde8 | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17) (MMPDE8). | |||||
|
PDE1C_HUMAN
|
||||||
| NC score | 0.903547 (rank : 21) | θ value | 6.59618e-35 (rank : 25) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
PDE1C_MOUSE
|
||||||
| NC score | 0.902329 (rank : 22) | θ value | 3.27365e-34 (rank : 26) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
PDE2A_MOUSE
|
||||||
| NC score | 0.901064 (rank : 23) | θ value | 4.88048e-38 (rank : 18) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q922S4, Q8K2U1 | Gene names | Pde2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE2A_HUMAN
|
||||||
| NC score | 0.900231 (rank : 24) | θ value | 4.88048e-38 (rank : 17) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00408 | Gene names | PDE2A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE11_HUMAN
|
||||||
| NC score | 0.889931 (rank : 25) | θ value | 3.37985e-39 (rank : 14) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HCR9, Q53T16, Q96S76, Q9GZY7, Q9HB46, Q9NY45 | Gene names | PDE11A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE11_MOUSE
|
||||||
| NC score | 0.889898 (rank : 26) | θ value | 8.89434e-40 (rank : 12) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P0C1Q2 | Gene names | Pde11a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE3B_HUMAN
|
||||||
| NC score | 0.884607 (rank : 27) | θ value | 2.96777e-27 (rank : 37) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13370, O00639, Q14408 | Gene names | PDE3B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase B (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase B) (CGI-PDE B) (CGIPDE1) (CGIP1). | |||||
|
PDE10_HUMAN
|
||||||
| NC score | 0.879796 (rank : 28) | θ value | 6.61148e-27 (rank : 38) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y233, Q9Y5T1 | Gene names | PDE10A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A (EC 3.1.4.17). | |||||
|
PDE3A_HUMAN
|
||||||
| NC score | 0.874453 (rank : 29) | θ value | 1.57365e-28 (rank : 34) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14432, O60865, Q13348 | Gene names | PDE3A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PDE3A_MOUSE
|
||||||
| NC score | 0.873256 (rank : 30) | θ value | 7.80994e-28 (rank : 36) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Z0X4 | Gene names | Pde3a | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PDE5A_MOUSE
|
||||||
| NC score | 0.871769 (rank : 31) | θ value | 2.34606e-32 (rank : 27) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CG03 | Gene names | Pde5a, Pde5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE5A_HUMAN
|
||||||
| NC score | 0.869579 (rank : 32) | θ value | 3.06405e-32 (rank : 28) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O76074, O75026, O75887, Q86UI0, Q86V66, Q9Y6Z6 | Gene names | PDE5A, PDE5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE6C_MOUSE
|
||||||
| NC score | 0.854186 (rank : 33) | θ value | 2.42779e-29 (rank : 33) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91ZQ1, Q8R0D4 | Gene names | Pde6c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE6C_HUMAN
|
||||||
| NC score | 0.853302 (rank : 34) | θ value | 1.08979e-29 (rank : 32) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P51160, Q5VY29 | Gene names | PDE6C, PDEA2 | |||
|
Domain Architecture |

|
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE6A_HUMAN
|
||||||
| NC score | 0.850441 (rank : 35) | θ value | 1.28732e-30 (rank : 30) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16499 | Gene names | PDE6A, PDEA | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha) (PDE V-B1). | |||||
|
PDE6B_HUMAN
|
||||||
| NC score | 0.848812 (rank : 36) | θ value | 1.28732e-30 (rank : 31) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35913, Q9BWH5 | Gene names | PDE6B, PDEB | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase beta-subunit precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PDE6B_MOUSE
|
||||||
| NC score | 0.847671 (rank : 37) | θ value | 1.16434e-31 (rank : 29) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P23440 | Gene names | Pde6b, Mpb, Pdeb, rd | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PDE6A_MOUSE
|
||||||
| NC score | 0.846012 (rank : 38) | θ value | 3.50572e-28 (rank : 35) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27664 | Gene names | Pde6a, Mpa, Pdea | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha). | |||||
|
SESN1_MOUSE
|
||||||
| NC score | 0.061199 (rank : 39) | θ value | 0.00134147 (rank : 39) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58006, Q7TNF3 | Gene names | Sesn1, Pa26, Sest1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sestrin-1 (p53-regulated protein PA26). | |||||
|
PDCD7_MOUSE
|
||||||
| NC score | 0.054919 (rank : 40) | θ value | 0.0330416 (rank : 41) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WTY1, Q8R5D9 | Gene names | Pdcd7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death protein 7 (ES18). | |||||
|
SESN1_HUMAN
|
||||||
| NC score | 0.053079 (rank : 41) | θ value | 0.0148317 (rank : 40) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6P5, Q9NV00, Q9UPD5, Q9Y6P6 | Gene names | SESN1, PA26, SEST1 | |||
|
Domain Architecture |

|
|||||
| Description | Sestrin-1 (p53-regulated protein PA26). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.039144 (rank : 42) | θ value | 0.0431538 (rank : 42) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.038961 (rank : 43) | θ value | 0.0736092 (rank : 45) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.038709 (rank : 44) | θ value | 0.0736092 (rank : 44) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.038377 (rank : 45) | θ value | 0.279714 (rank : 47) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.038204 (rank : 46) | θ value | 0.0563607 (rank : 43) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.037698 (rank : 47) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
PARC_HUMAN
|
||||||
| NC score | 0.037135 (rank : 48) | θ value | 0.0736092 (rank : 46) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.036697 (rank : 49) | θ value | 0.279714 (rank : 48) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.036120 (rank : 50) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.034163 (rank : 51) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.034101 (rank : 52) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.033789 (rank : 53) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
PDCD7_HUMAN
|
||||||
| NC score | 0.033366 (rank : 54) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N8D1, Q96AK8, Q9Y6D7 | Gene names | PDCD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death protein 7 (ES18) (HES18). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.033227 (rank : 55) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.031939 (rank : 56) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.031605 (rank : 57) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
PARC_MOUSE
|
||||||
| NC score | 0.030997 (rank : 58) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80TT8, Q8BKL9, Q8CGC0, Q8R363 | Gene names | Parc, Kiaa0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein. | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.029893 (rank : 59) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.029271 (rank : 60) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.028956 (rank : 61) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.028873 (rank : 62) | θ value | 0.47712 (rank : 49) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
BRE1B_MOUSE
|
||||||
| NC score | 0.027058 (rank : 63) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
SMC4_HUMAN
|
||||||
| NC score | 0.027022 (rank : 64) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
BRE1B_HUMAN
|
||||||
| NC score | 0.026060 (rank : 65) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.025808 (rank : 66) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.025595 (rank : 67) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
TMF1_HUMAN
|
||||||
| NC score | 0.025413 (rank : 68) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.024103 (rank : 69) | θ value | 0.62314 (rank : 51) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
SM1L2_MOUSE
|
||||||
| NC score | 0.023909 (rank : 70) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
SM1L2_HUMAN
|
||||||
| NC score | 0.022935 (rank : 71) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.022404 (rank : 72) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.021983 (rank : 73) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.021145 (rank : 74) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
LZTS1_MOUSE
|
||||||
| NC score | 0.020468 (rank : 75) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P60853 | Gene names | Lzts1, Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/Esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.019817 (rank : 76) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.018993 (rank : 77) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.018894 (rank : 78) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.017238 (rank : 79) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
RADI_HUMAN
|
||||||
| NC score | 0.016916 (rank : 80) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.016849 (rank : 81) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
RADI_MOUSE
|
||||||
| NC score | 0.014008 (rank : 82) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.008927 (rank : 83) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
JPH3_MOUSE
|
||||||
| NC score | 0.007104 (rank : 84) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ET77, Q9EQZ2 | Gene names | Jph3, Jp3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
DESM_HUMAN
|
||||||
| NC score | 0.006961 (rank : 85) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P17661, Q15787, Q8IZR1, Q8IZR6, Q8NES2, Q8NEU6, Q8TAC4, Q8TCX2, Q8TD99, Q9UHN5, Q9UJ80 | Gene names | DES | |||
|
Domain Architecture |

|
|||||
| Description | Desmin. | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.006943 (rank : 86) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
PIAS1_HUMAN
|
||||||
| NC score | 0.006752 (rank : 87) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75925, Q99751, Q9UN02 | Gene names | PIAS1, DDXBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 1 (Gu-binding protein) (GBP) (RNA helicase II-binding protein) (DEAD/H box-binding protein 1). | |||||
|
2A5A_MOUSE
|
||||||
| NC score | 0.004129 (rank : 88) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PD03, Q8R1U7 | Gene names | Ppp2r5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform (PP2A, B subunit, B' alpha isoform) (PP2A, B subunit, B56 alpha isoform) (PP2A, B subunit, PR61 alpha isoform) (PP2A, B subunit, R5 alpha isoform) (PR61alpha). | |||||
|
E41L2_MOUSE
|
||||||
| NC score | 0.004000 (rank : 89) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
ANR47_MOUSE
|
||||||
| NC score | 0.002642 (rank : 90) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z1P7 | Gene names | Ankrd47, D17Ertd288e, Ng28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 47. | |||||
|
SPAST_MOUSE
|
||||||
| NC score | 0.001796 (rank : 91) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QYY8, Q6ZPY6, Q80VE0, Q9CVK0 | Gene names | Spast, Kiaa1083, Spg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spastin. | |||||
|
CD2L1_MOUSE
|
||||||
| NC score | -0.002737 (rank : 92) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | -0.004046 (rank : 93) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||