Please be patient as the page loads
|
PDE3A_HUMAN
|
||||||
| SwissProt Accessions | Q14432, O60865, Q13348 | Gene names | PDE3A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PDE3A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q14432, O60865, Q13348 | Gene names | PDE3A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PDE3A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.993518 (rank : 2) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Z0X4 | Gene names | Pde3a | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PDE3B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.986246 (rank : 3) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13370, O00639, Q14408 | Gene names | PDE3B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase B (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase B) (CGI-PDE B) (CGIPDE1) (CGIP1). | |||||
|
PDE1C_MOUSE
|
||||||
| θ value | 1.51363e-47 (rank : 4) | NC score | 0.903157 (rank : 9) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
PDE1A_HUMAN
|
||||||
| θ value | 1.97686e-47 (rank : 5) | NC score | 0.912191 (rank : 5) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P54750, Q86VZ0, Q9C0K8, Q9C0K9, Q9C0L0, Q9C0L1, Q9C0L2, Q9C0L3, Q9C0L4, Q9UFX3 | Gene names | PDE1A | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE) (hCam-1). | |||||
|
PDE1A_MOUSE
|
||||||
| θ value | 1.97686e-47 (rank : 6) | NC score | 0.912732 (rank : 4) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61481, O35388 | Gene names | Pde1a | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE). | |||||
|
PDE1C_HUMAN
|
||||||
| θ value | 2.85459e-46 (rank : 7) | NC score | 0.903226 (rank : 8) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
PDE4D_MOUSE
|
||||||
| θ value | 1.73182e-43 (rank : 8) | NC score | 0.892901 (rank : 13) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01063, Q6TRH9, Q8C4Q7, Q8CG05 | Gene names | Pde4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3). | |||||
|
PDE4D_HUMAN
|
||||||
| θ value | 3.85808e-43 (rank : 9) | NC score | 0.891950 (rank : 15) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08499, O43433, Q13549, Q13550, Q13551, Q7Z2L8, Q8IV84, Q8IVA9, Q8IVD2, Q8IVD3, Q9HCX7 | Gene names | PDE4D | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3) (PDE43). | |||||
|
PDE4A_MOUSE
|
||||||
| θ value | 6.58091e-43 (rank : 10) | NC score | 0.893722 (rank : 12) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O89084, Q8R078, Q9JHQ4, Q9QX48, Q9QX49, Q9QXI8 | Gene names | Pde4a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17). | |||||
|
PDE4A_HUMAN
|
||||||
| θ value | 1.91475e-42 (rank : 11) | NC score | 0.892752 (rank : 14) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27815, O75522, O76092, Q16255, Q16691, Q8WUQ3 | Gene names | PDE4A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17) (DPDE2) (PDE46). | |||||
|
PDE4B_HUMAN
|
||||||
| θ value | 1.91475e-42 (rank : 12) | NC score | 0.894942 (rank : 10) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q07343, O15443 | Gene names | PDE4B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4B (EC 3.1.4.17) (DPDE4) (PDE32). | |||||
|
PDE1B_MOUSE
|
||||||
| θ value | 8.04462e-41 (rank : 13) | NC score | 0.908470 (rank : 6) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01065, O35384 | Gene names | Pde1b, Pde1b1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE1B_HUMAN
|
||||||
| θ value | 1.79215e-40 (rank : 14) | NC score | 0.907457 (rank : 7) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01064, Q92825 | Gene names | PDE1B, PDE1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE4C_MOUSE
|
||||||
| θ value | 7.52953e-39 (rank : 15) | NC score | 0.894584 (rank : 11) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3UEI1, Q8K0P4 | Gene names | Pde4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17). | |||||
|
PDE4C_HUMAN
|
||||||
| θ value | 1.6774e-38 (rank : 16) | NC score | 0.891430 (rank : 16) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q08493, Q9UN44, Q9UN45, Q9UN46, Q9UPJ6 | Gene names | PDE4C | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17) (DPDE1) (PDE21). | |||||
|
PDE8B_HUMAN
|
||||||
| θ value | 3.4976e-36 (rank : 17) | NC score | 0.888103 (rank : 17) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95263, Q86XK8, Q8IUJ7, Q8IUJ8, Q8IUJ9, Q8IUK0, Q8N3T2 | Gene names | PDE8B | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8B (EC 3.1.4.17) (HSPDE8B). | |||||
|
PDE8A_MOUSE
|
||||||
| θ value | 6.59618e-35 (rank : 18) | NC score | 0.885095 (rank : 20) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88502 | Gene names | Pde8a, Pde8 | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17) (MMPDE8). | |||||
|
PDE8A_HUMAN
|
||||||
| θ value | 3.27365e-34 (rank : 19) | NC score | 0.885040 (rank : 21) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O60658, Q969I1, Q96PC9, Q96PD0, Q96PD1, Q9UMB7 | Gene names | PDE8A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17). | |||||
|
PDE7A_HUMAN
|
||||||
| θ value | 2.19584e-30 (rank : 20) | NC score | 0.886066 (rank : 19) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13946, O15380 | Gene names | PDE7A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (HCP1) (TM22). | |||||
|
PDE7A_MOUSE
|
||||||
| θ value | 3.74554e-30 (rank : 21) | NC score | 0.886794 (rank : 18) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70453, Q9ERB3 | Gene names | Pde7a | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (P2A). | |||||
|
PDE9A_HUMAN
|
||||||
| θ value | 1.57365e-28 (rank : 22) | NC score | 0.874453 (rank : 25) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O76083, O75490, O75491, O95225, Q86SF7, Q86SI6, Q86SJ3, Q86WN3, Q86WN4, Q86WN5, Q86WN6, Q86WN7, Q86WN8, Q86WN9, Q86WP0 | Gene names | PDE9A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE9A_MOUSE
|
||||||
| θ value | 2.05525e-28 (rank : 23) | NC score | 0.878117 (rank : 24) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O70628 | Gene names | Pde9a, Pde8b | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE7B_MOUSE
|
||||||
| θ value | 8.63488e-27 (rank : 24) | NC score | 0.881010 (rank : 22) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QXQ1 | Gene names | Pde7b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE7B_HUMAN
|
||||||
| θ value | 1.62847e-25 (rank : 25) | NC score | 0.878797 (rank : 23) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NP56 | Gene names | PDE7B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE2A_HUMAN
|
||||||
| θ value | 1.02238e-19 (rank : 26) | NC score | 0.791781 (rank : 27) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00408 | Gene names | PDE2A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE2A_MOUSE
|
||||||
| θ value | 1.02238e-19 (rank : 27) | NC score | 0.792778 (rank : 26) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q922S4, Q8K2U1 | Gene names | Pde2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE11_HUMAN
|
||||||
| θ value | 8.65492e-19 (rank : 28) | NC score | 0.765620 (rank : 28) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HCR9, Q53T16, Q96S76, Q9GZY7, Q9HB46, Q9NY45 | Gene names | PDE11A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE11_MOUSE
|
||||||
| θ value | 1.92812e-18 (rank : 29) | NC score | 0.764956 (rank : 29) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P0C1Q2 | Gene names | Pde11a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE6C_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 30) | NC score | 0.730039 (rank : 34) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P51160, Q5VY29 | Gene names | PDE6C, PDEA2 | |||
|
Domain Architecture |

|
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE6C_MOUSE
|
||||||
| θ value | 8.95645e-16 (rank : 31) | NC score | 0.730291 (rank : 33) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91ZQ1, Q8R0D4 | Gene names | Pde6c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE6A_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 32) | NC score | 0.723404 (rank : 35) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16499 | Gene names | PDE6A, PDEA | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha) (PDE V-B1). | |||||
|
PDE6A_MOUSE
|
||||||
| θ value | 4.91457e-14 (rank : 33) | NC score | 0.719786 (rank : 36) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27664 | Gene names | Pde6a, Mpa, Pdea | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha). | |||||
|
PDE6B_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 34) | NC score | 0.719321 (rank : 37) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35913, Q9BWH5 | Gene names | PDE6B, PDEB | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase beta-subunit precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PDE6B_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 35) | NC score | 0.714793 (rank : 38) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P23440 | Gene names | Pde6b, Mpb, Pdeb, rd | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PDE10_HUMAN
|
||||||
| θ value | 2.98157e-11 (rank : 36) | NC score | 0.750970 (rank : 30) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y233, Q9Y5T1 | Gene names | PDE10A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A (EC 3.1.4.17). | |||||
|
PDE5A_HUMAN
|
||||||
| θ value | 2.98157e-11 (rank : 37) | NC score | 0.731002 (rank : 32) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O76074, O75026, O75887, Q86UI0, Q86V66, Q9Y6Z6 | Gene names | PDE5A, PDE5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE5A_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 38) | NC score | 0.732189 (rank : 31) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CG03 | Gene names | Pde5a, Pde5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
KI18A_HUMAN
|
||||||
| θ value | 0.163984 (rank : 39) | NC score | 0.009098 (rank : 53) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NI77, Q86VS5, Q9H0F3 | Gene names | KIF18A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 18A. | |||||
|
IRF2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 40) | NC score | 0.023310 (rank : 39) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23906 | Gene names | Irf2 | |||
|
Domain Architecture |
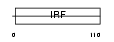
|
|||||
| Description | Interferon regulatory factor 2 (IRF-2). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.016841 (rank : 44) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
ANGP2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.008207 (rank : 55) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35608, Q3U1A1, Q9D2D2 | Gene names | Angpt2, Agpt2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-2 precursor (ANG-2). | |||||
|
MYO3A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | -0.002345 (rank : 78) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEV4, Q8WX17, Q9NYS8 | Gene names | MYO3A | |||
|
Domain Architecture |

|
|||||
| Description | Myosin IIIA (EC 2.7.11.1). | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.016665 (rank : 45) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
DCP1A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.014503 (rank : 49) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YD3, Q6NZE3 | Gene names | Dcp1a, Mitc1, Smif | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (MAD homolog 4-interacting transcription coactivator 1) (Smad4-interacting transcriptional co-activator). | |||||
|
HASP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.016126 (rank : 47) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0R0, Q99MU9, Q9JJJ4 | Gene names | Gsg2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Haspin (EC 2.7.11.1) (Haploid germ cell-specific nuclear protein kinase) (Germ cell-specific gene 2 protein). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.010970 (rank : 52) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LEUK_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.016115 (rank : 48) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P16150 | Gene names | SPN, CD43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Galactoglycoprotein) (GALGP) (CD43 antigen). | |||||
|
PAX3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.003317 (rank : 67) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P24610, Q3UFQ9, Q8BRE7, Q9CXI6 | Gene names | Pax3, Pax-3 | |||
|
Domain Architecture |
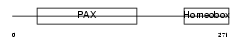
|
|||||
| Description | Paired box protein Pax-3. | |||||
|
COLI_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.022303 (rank : 41) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01193, P01200, Q544U4 | Gene names | Pomc, Pomc1 | |||
|
Domain Architecture |
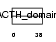
|
|||||
| Description | Corticotropin-lipotropin precursor (Pro-opiomelanocortin) (POMC) [Contains: NPP; Melanotropin gamma (Gamma-MSH); Corticotropin (Adrenocorticotropic hormone) (ACTH); Melanotropin alpha (Alpha-MSH); Corticotropin-like intermediary peptide (CLIP); Lipotropin beta (Beta- LPH); Lipotropin gamma (Gamma-LPH); Melanotropin beta (Beta-MSH); Beta-endorphin; Met-enkephalin]. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.001252 (rank : 72) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
MAPK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | -0.000705 (rank : 75) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49137, Q8IYD6 | Gene names | MAPKAPK2 | |||
|
Domain Architecture |
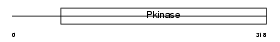
|
|||||
| Description | MAP kinase-activated protein kinase 2 (EC 2.7.11.1) (MAPK-activated protein kinase 2) (MAPKAP kinase 2) (MAPKAPK-2) (MK2). | |||||
|
MAPK2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | -0.000682 (rank : 74) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49138, Q6P561 | Gene names | Mapkapk2, Rps6kc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAP kinase-activated protein kinase 2 (EC 2.7.11.1) (MAPK-activated protein kinase 2) (MAPKAP kinase 2) (MAPKAPK-2) (MK2). | |||||
|
CAD26_MOUSE
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.002042 (rank : 70) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59862 | Gene names | Cdh26 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-like protein 26 precursor. | |||||
|
CN113_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.022341 (rank : 40) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NXV8 | Gene names | C14orf113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf113. | |||||
|
IRF2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.016539 (rank : 46) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14316, Q96B99 | Gene names | IRF2 | |||
|
Domain Architecture |
|
|||||
| Description | Interferon regulatory factor 2 (IRF-2). | |||||
|
PAX3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.002739 (rank : 68) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23760, Q16448 | Gene names | PAX3, HUP2 | |||
|
Domain Architecture |
|
|||||
| Description | Paired box protein Pax-3 (HUP2). | |||||
|
DONS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.011394 (rank : 51) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXP4, Q80V93, Q8BV55 | Gene names | Donson, ORF60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein downstream neighbor of Son (Protein 3SG). | |||||
|
GLIS3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | -0.003657 (rank : 79) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 758 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEA6 | Gene names | GLIS3, ZNF515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3) (Zinc finger protein 515). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.013470 (rank : 50) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.017228 (rank : 43) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ATX2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.008486 (rank : 54) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
COLI_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.017683 (rank : 42) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01189, P78442, Q9UD39, Q9UD40 | Gene names | POMC | |||
|
Domain Architecture |

|
|||||
| Description | Corticotropin-lipotropin precursor (Pro-opiomelanocortin) (POMC) [Contains: NPP; Melanotropin gamma (Gamma-MSH); Potential peptide; Corticotropin (Adrenocorticotropic hormone) (ACTH); Melanotropin alpha (Alpha-MSH); Corticotropin-like intermediary peptide (CLIP); Lipotropin beta (Beta-LPH); Lipotropin gamma (Gamma-LPH); Melanotropin beta (Beta-MSH); Beta-endorphin; Met-enkephalin]. | |||||
|
FXL19_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.007494 (rank : 56) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PB97, Q7TSH0, Q8BIB4 | Gene names | Fbxl19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 19 (F-box and leucine-rich repeat protein 19). | |||||
|
FZD5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.003682 (rank : 65) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13467, Q53R22 | Gene names | FZD5, HFZ5 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-5 precursor (Fz-5) (hFz5) (FzE5). | |||||
|
FZD5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.003695 (rank : 64) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQD0, O08975, Q8BMR2, Q8CHK9 | Gene names | Fzd5 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-5 precursor (Fz-5) (mFz5). | |||||
|
KCNB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.001373 (rank : 71) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
PTPRT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.000356 (rank : 73) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99M80, Q99M81, Q99M82, Q9JIZ1, Q9JIZ2, Q9JKC2, Q9JLP0 | Gene names | Ptprt | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho) (mRPTPrho) (RPTPmam4). | |||||
|
SP100_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.004543 (rank : 62) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23497, O75450, Q13343, Q96F70, Q96T24, Q96T95, Q9NP33, Q9UE32 | Gene names | SP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein) (Lysp100b). | |||||
|
ANKR6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | -0.000956 (rank : 77) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
CA103_MOUSE
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.007298 (rank : 57) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CDD9, Q8C893 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C1orf103 homolog. | |||||
|
GPR97_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.005422 (rank : 60) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R0T6 | Gene names | Gpr97 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 97 precursor. | |||||
|
KI67_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.005820 (rank : 59) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
LIMA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.003468 (rank : 66) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERG0, Q9ERG1 | Gene names | Lima1, D15Ertd366e, Eplin | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm) (mEPLIN). | |||||
|
LRRF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | -0.000955 (rank : 76) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
SETBP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.006073 (rank : 58) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
SVIL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.004238 (rank : 63) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95425, O60611, O60612, Q5VZK5, Q5VZK6, Q9H1R7 | Gene names | SVIL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.005254 (rank : 61) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TGM3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.002733 (rank : 69) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08189 | Gene names | Tgm3, Tgase3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase E precursor (EC 2.3.2.13) (TGase E) (TGE) (TG(E)) (Transglutaminase-3) [Contains: Protein- glutamine gamma-glutamyltransferase E 50 kDa non-catalytic chain; Protein-glutamine gamma-glutamyltransferase E 27 kDa catalytic chain]. | |||||
|
PDE3A_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q14432, O60865, Q13348 | Gene names | PDE3A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PDE3A_MOUSE
|
||||||
| NC score | 0.993518 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Z0X4 | Gene names | Pde3a | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PDE3B_HUMAN
|
||||||
| NC score | 0.986246 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13370, O00639, Q14408 | Gene names | PDE3B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase B (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase B) (CGI-PDE B) (CGIPDE1) (CGIP1). | |||||
|
PDE1A_MOUSE
|
||||||
| NC score | 0.912732 (rank : 4) | θ value | 1.97686e-47 (rank : 6) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61481, O35388 | Gene names | Pde1a | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE). | |||||
|
PDE1A_HUMAN
|
||||||
| NC score | 0.912191 (rank : 5) | θ value | 1.97686e-47 (rank : 5) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P54750, Q86VZ0, Q9C0K8, Q9C0K9, Q9C0L0, Q9C0L1, Q9C0L2, Q9C0L3, Q9C0L4, Q9UFX3 | Gene names | PDE1A | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE) (hCam-1). | |||||
|
PDE1B_MOUSE
|
||||||
| NC score | 0.908470 (rank : 6) | θ value | 8.04462e-41 (rank : 13) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01065, O35384 | Gene names | Pde1b, Pde1b1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE1B_HUMAN
|
||||||
| NC score | 0.907457 (rank : 7) | θ value | 1.79215e-40 (rank : 14) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01064, Q92825 | Gene names | PDE1B, PDE1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE1C_HUMAN
|
||||||
| NC score | 0.903226 (rank : 8) | θ value | 2.85459e-46 (rank : 7) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
PDE1C_MOUSE
|
||||||
| NC score | 0.903157 (rank : 9) | θ value | 1.51363e-47 (rank : 4) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
PDE4B_HUMAN
|
||||||
| NC score | 0.894942 (rank : 10) | θ value | 1.91475e-42 (rank : 12) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q07343, O15443 | Gene names | PDE4B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4B (EC 3.1.4.17) (DPDE4) (PDE32). | |||||
|
PDE4C_MOUSE
|
||||||
| NC score | 0.894584 (rank : 11) | θ value | 7.52953e-39 (rank : 15) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3UEI1, Q8K0P4 | Gene names | Pde4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17). | |||||
|
PDE4A_MOUSE
|
||||||
| NC score | 0.893722 (rank : 12) | θ value | 6.58091e-43 (rank : 10) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O89084, Q8R078, Q9JHQ4, Q9QX48, Q9QX49, Q9QXI8 | Gene names | Pde4a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17). | |||||
|
PDE4D_MOUSE
|
||||||
| NC score | 0.892901 (rank : 13) | θ value | 1.73182e-43 (rank : 8) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01063, Q6TRH9, Q8C4Q7, Q8CG05 | Gene names | Pde4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3). | |||||
|
PDE4A_HUMAN
|
||||||
| NC score | 0.892752 (rank : 14) | θ value | 1.91475e-42 (rank : 11) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27815, O75522, O76092, Q16255, Q16691, Q8WUQ3 | Gene names | PDE4A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17) (DPDE2) (PDE46). | |||||
|
PDE4D_HUMAN
|
||||||
| NC score | 0.891950 (rank : 15) | θ value | 3.85808e-43 (rank : 9) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08499, O43433, Q13549, Q13550, Q13551, Q7Z2L8, Q8IV84, Q8IVA9, Q8IVD2, Q8IVD3, Q9HCX7 | Gene names | PDE4D | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3) (PDE43). | |||||
|
PDE4C_HUMAN
|
||||||
| NC score | 0.891430 (rank : 16) | θ value | 1.6774e-38 (rank : 16) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q08493, Q9UN44, Q9UN45, Q9UN46, Q9UPJ6 | Gene names | PDE4C | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17) (DPDE1) (PDE21). | |||||
|
PDE8B_HUMAN
|
||||||
| NC score | 0.888103 (rank : 17) | θ value | 3.4976e-36 (rank : 17) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95263, Q86XK8, Q8IUJ7, Q8IUJ8, Q8IUJ9, Q8IUK0, Q8N3T2 | Gene names | PDE8B | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8B (EC 3.1.4.17) (HSPDE8B). | |||||
|
PDE7A_MOUSE
|
||||||
| NC score | 0.886794 (rank : 18) | θ value | 3.74554e-30 (rank : 21) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70453, Q9ERB3 | Gene names | Pde7a | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (P2A). | |||||
|
PDE7A_HUMAN
|
||||||
| NC score | 0.886066 (rank : 19) | θ value | 2.19584e-30 (rank : 20) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13946, O15380 | Gene names | PDE7A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (HCP1) (TM22). | |||||
|
PDE8A_MOUSE
|
||||||
| NC score | 0.885095 (rank : 20) | θ value | 6.59618e-35 (rank : 18) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88502 | Gene names | Pde8a, Pde8 | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17) (MMPDE8). | |||||
|
PDE8A_HUMAN
|
||||||
| NC score | 0.885040 (rank : 21) | θ value | 3.27365e-34 (rank : 19) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O60658, Q969I1, Q96PC9, Q96PD0, Q96PD1, Q9UMB7 | Gene names | PDE8A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17). | |||||
|
PDE7B_MOUSE
|
||||||
| NC score | 0.881010 (rank : 22) | θ value | 8.63488e-27 (rank : 24) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QXQ1 | Gene names | Pde7b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE7B_HUMAN
|
||||||
| NC score | 0.878797 (rank : 23) | θ value | 1.62847e-25 (rank : 25) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NP56 | Gene names | PDE7B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE9A_MOUSE
|
||||||
| NC score | 0.878117 (rank : 24) | θ value | 2.05525e-28 (rank : 23) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O70628 | Gene names | Pde9a, Pde8b | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE9A_HUMAN
|
||||||
| NC score | 0.874453 (rank : 25) | θ value | 1.57365e-28 (rank : 22) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O76083, O75490, O75491, O95225, Q86SF7, Q86SI6, Q86SJ3, Q86WN3, Q86WN4, Q86WN5, Q86WN6, Q86WN7, Q86WN8, Q86WN9, Q86WP0 | Gene names | PDE9A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE2A_MOUSE
|
||||||
| NC score | 0.792778 (rank : 26) | θ value | 1.02238e-19 (rank : 27) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q922S4, Q8K2U1 | Gene names | Pde2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE2A_HUMAN
|
||||||
| NC score | 0.791781 (rank : 27) | θ value | 1.02238e-19 (rank : 26) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00408 | Gene names | PDE2A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE11_HUMAN
|
||||||
| NC score | 0.765620 (rank : 28) | θ value | 8.65492e-19 (rank : 28) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HCR9, Q53T16, Q96S76, Q9GZY7, Q9HB46, Q9NY45 | Gene names | PDE11A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE11_MOUSE
|
||||||
| NC score | 0.764956 (rank : 29) | θ value | 1.92812e-18 (rank : 29) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P0C1Q2 | Gene names | Pde11a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE10_HUMAN
|
||||||
| NC score | 0.750970 (rank : 30) | θ value | 2.98157e-11 (rank : 36) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y233, Q9Y5T1 | Gene names | PDE10A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A (EC 3.1.4.17). | |||||
|
PDE5A_MOUSE
|
||||||
| NC score | 0.732189 (rank : 31) | θ value | 2.52405e-10 (rank : 38) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CG03 | Gene names | Pde5a, Pde5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE5A_HUMAN
|
||||||
| NC score | 0.731002 (rank : 32) | θ value | 2.98157e-11 (rank : 37) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O76074, O75026, O75887, Q86UI0, Q86V66, Q9Y6Z6 | Gene names | PDE5A, PDE5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE6C_MOUSE
|
||||||
| NC score | 0.730291 (rank : 33) | θ value | 8.95645e-16 (rank : 31) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91ZQ1, Q8R0D4 | Gene names | Pde6c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE6C_HUMAN
|
||||||
| NC score | 0.730039 (rank : 34) | θ value | 4.02038e-16 (rank : 30) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P51160, Q5VY29 | Gene names | PDE6C, PDEA2 | |||
|
Domain Architecture |

|
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE6A_HUMAN
|
||||||
| NC score | 0.723404 (rank : 35) | θ value | 2.20605e-14 (rank : 32) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16499 | Gene names | PDE6A, PDEA | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha) (PDE V-B1). | |||||
|
PDE6A_MOUSE
|
||||||
| NC score | 0.719786 (rank : 36) | θ value | 4.91457e-14 (rank : 33) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27664 | Gene names | Pde6a, Mpa, Pdea | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha). | |||||
|
PDE6B_HUMAN
|
||||||
| NC score | 0.719321 (rank : 37) | θ value | 1.86753e-13 (rank : 34) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35913, Q9BWH5 | Gene names | PDE6B, PDEB | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase beta-subunit precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PDE6B_MOUSE
|
||||||
| NC score | 0.714793 (rank : 38) | θ value | 3.52202e-12 (rank : 35) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P23440 | Gene names | Pde6b, Mpb, Pdeb, rd | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
IRF2_MOUSE
|
||||||
| NC score | 0.023310 (rank : 39) | θ value | 0.21417 (rank : 40) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23906 | Gene names | Irf2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 2 (IRF-2). | |||||
|
CN113_HUMAN
|
||||||
| NC score | 0.022341 (rank : 40) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NXV8 | Gene names | C14orf113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf113. | |||||
|
COLI_MOUSE
|
||||||
| NC score | 0.022303 (rank : 41) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01193, P01200, Q544U4 | Gene names | Pomc, Pomc1 | |||
|
Domain Architecture |

|
|||||
| Description | Corticotropin-lipotropin precursor (Pro-opiomelanocortin) (POMC) [Contains: NPP; Melanotropin gamma (Gamma-MSH); Corticotropin (Adrenocorticotropic hormone) (ACTH); Melanotropin alpha (Alpha-MSH); Corticotropin-like intermediary peptide (CLIP); Lipotropin beta (Beta- LPH); Lipotropin gamma (Gamma-LPH); Melanotropin beta (Beta-MSH); Beta-endorphin; Met-enkephalin]. | |||||
|
COLI_HUMAN
|
||||||
| NC score | 0.017683 (rank : 42) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01189, P78442, Q9UD39, Q9UD40 | Gene names | POMC | |||
|
Domain Architecture |

|
|||||
| Description | Corticotropin-lipotropin precursor (Pro-opiomelanocortin) (POMC) [Contains: NPP; Melanotropin gamma (Gamma-MSH); Potential peptide; Corticotropin (Adrenocorticotropic hormone) (ACTH); Melanotropin alpha (Alpha-MSH); Corticotropin-like intermediary peptide (CLIP); Lipotropin beta (Beta-LPH); Lipotropin gamma (Gamma-LPH); Melanotropin beta (Beta-MSH); Beta-endorphin; Met-enkephalin]. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.017228 (rank : 43) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.016841 (rank : 44) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.016665 (rank : 45) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
IRF2_HUMAN
|
||||||
| NC score | 0.016539 (rank : 46) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14316, Q96B99 | Gene names | IRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 2 (IRF-2). | |||||
|
HASP_MOUSE
|
||||||
| NC score | 0.016126 (rank : 47) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0R0, Q99MU9, Q9JJJ4 | Gene names | Gsg2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Haspin (EC 2.7.11.1) (Haploid germ cell-specific nuclear protein kinase) (Germ cell-specific gene 2 protein). | |||||
|
LEUK_HUMAN
|
||||||
| NC score | 0.016115 (rank : 48) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P16150 | Gene names | SPN, CD43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Galactoglycoprotein) (GALGP) (CD43 antigen). | |||||
|
DCP1A_MOUSE
|
||||||
| NC score | 0.014503 (rank : 49) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YD3, Q6NZE3 | Gene names | Dcp1a, Mitc1, Smif | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (MAD homolog 4-interacting transcription coactivator 1) (Smad4-interacting transcriptional co-activator). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.013470 (rank : 50) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
DONS_MOUSE
|
||||||
| NC score | 0.011394 (rank : 51) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXP4, Q80V93, Q8BV55 | Gene names | Donson, ORF60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein downstream neighbor of Son (Protein 3SG). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.010970 (rank : 52) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
KI18A_HUMAN
|
||||||
| NC score | 0.009098 (rank : 53) | θ value | 0.163984 (rank : 39) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NI77, Q86VS5, Q9H0F3 | Gene names | KIF18A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 18A. | |||||
|
ATX2_HUMAN
|
||||||
| NC score | 0.008486 (rank : 54) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
ANGP2_MOUSE
|
||||||
| NC score | 0.008207 (rank : 55) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35608, Q3U1A1, Q9D2D2 | Gene names | Angpt2, Agpt2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-2 precursor (ANG-2). | |||||
|
FXL19_MOUSE
|
||||||
| NC score | 0.007494 (rank : 56) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PB97, Q7TSH0, Q8BIB4 | Gene names | Fbxl19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 19 (F-box and leucine-rich repeat protein 19). | |||||
|
CA103_MOUSE
|
||||||
| NC score | 0.007298 (rank : 57) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CDD9, Q8C893 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C1orf103 homolog. | |||||
|
SETBP_HUMAN
|
||||||
| NC score | 0.006073 (rank : 58) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.005820 (rank : 59) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
GPR97_MOUSE
|
||||||
| NC score | 0.005422 (rank : 60) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R0T6 | Gene names | Gpr97 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 97 precursor. | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.005254 (rank : 61) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
SP100_HUMAN
|
||||||
| NC score | 0.004543 (rank : 62) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23497, O75450, Q13343, Q96F70, Q96T24, Q96T95, Q9NP33, Q9UE32 | Gene names | SP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein) (Lysp100b). | |||||
|
SVIL_HUMAN
|
||||||
| NC score | 0.004238 (rank : 63) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95425, O60611, O60612, Q5VZK5, Q5VZK6, Q9H1R7 | Gene names | SVIL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
FZD5_MOUSE
|
||||||
| NC score | 0.003695 (rank : 64) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQD0, O08975, Q8BMR2, Q8CHK9 | Gene names | Fzd5 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-5 precursor (Fz-5) (mFz5). | |||||
|
FZD5_HUMAN
|
||||||
| NC score | 0.003682 (rank : 65) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13467, Q53R22 | Gene names | FZD5, HFZ5 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-5 precursor (Fz-5) (hFz5) (FzE5). | |||||
|
LIMA1_MOUSE
|
||||||
| NC score | 0.003468 (rank : 66) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERG0, Q9ERG1 | Gene names | Lima1, D15Ertd366e, Eplin | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm) (mEPLIN). | |||||
|
PAX3_MOUSE
|
||||||
| NC score | 0.003317 (rank : 67) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P24610, Q3UFQ9, Q8BRE7, Q9CXI6 | Gene names | Pax3, Pax-3 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-3. | |||||
|
PAX3_HUMAN
|
||||||
| NC score | 0.002739 (rank : 68) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23760, Q16448 | Gene names | PAX3, HUP2 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-3 (HUP2). | |||||
|
TGM3_MOUSE
|
||||||
| NC score | 0.002733 (rank : 69) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08189 | Gene names | Tgm3, Tgase3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase E precursor (EC 2.3.2.13) (TGase E) (TGE) (TG(E)) (Transglutaminase-3) [Contains: Protein- glutamine gamma-glutamyltransferase E 50 kDa non-catalytic chain; Protein-glutamine gamma-glutamyltransferase E 27 kDa catalytic chain]. | |||||
|
CAD26_MOUSE
|
||||||
| NC score | 0.002042 (rank : 70) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59862 | Gene names | Cdh26 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-like protein 26 precursor. | |||||
|
KCNB1_HUMAN
|
||||||
| NC score | 0.001373 (rank : 71) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.001252 (rank : 72) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
PTPRT_MOUSE
|
||||||
| NC score | 0.000356 (rank : 73) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99M80, Q99M81, Q99M82, Q9JIZ1, Q9JIZ2, Q9JKC2, Q9JLP0 | Gene names | Ptprt | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho) (mRPTPrho) (RPTPmam4). | |||||
|
MAPK2_MOUSE
|
||||||
| NC score | -0.000682 (rank : 74) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49138, Q6P561 | Gene names | Mapkapk2, Rps6kc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAP kinase-activated protein kinase 2 (EC 2.7.11.1) (MAPK-activated protein kinase 2) (MAPKAP kinase 2) (MAPKAPK-2) (MK2). | |||||
|
MAPK2_HUMAN
|
||||||
| NC score | -0.000705 (rank : 75) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49137, Q8IYD6 | Gene names | MAPKAPK2 | |||
|
Domain Architecture |

|
|||||
| Description | MAP kinase-activated protein kinase 2 (EC 2.7.11.1) (MAPK-activated protein kinase 2) (MAPKAP kinase 2) (MAPKAPK-2) (MK2). | |||||
|
LRRF2_HUMAN
|
||||||
| NC score | -0.000955 (rank : 76) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
ANKR6_HUMAN
|
||||||
| NC score | -0.000956 (rank : 77) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
MYO3A_HUMAN
|
||||||
| NC score | -0.002345 (rank : 78) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEV4, Q8WX17, Q9NYS8 | Gene names | MYO3A | |||
|
Domain Architecture |

|
|||||
| Description | Myosin IIIA (EC 2.7.11.1). | |||||
|
GLIS3_HUMAN
|
||||||
| NC score | -0.003657 (rank : 79) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 758 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEA6 | Gene names | GLIS3, ZNF515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3) (Zinc finger protein 515). | |||||