Please be patient as the page loads
|
PDE9A_MOUSE
|
||||||
| SwissProt Accessions | O70628 | Gene names | Pde9a, Pde8b | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PDE9A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.997194 (rank : 2) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O76083, O75490, O75491, O95225, Q86SF7, Q86SI6, Q86SJ3, Q86WN3, Q86WN4, Q86WN5, Q86WN6, Q86WN7, Q86WN8, Q86WN9, Q86WP0 | Gene names | PDE9A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE9A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O70628 | Gene names | Pde9a, Pde8b | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE4B_HUMAN
|
||||||
| θ value | 1.41672e-45 (rank : 3) | NC score | 0.924204 (rank : 9) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q07343, O15443 | Gene names | PDE4B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4B (EC 3.1.4.17) (DPDE4) (PDE32). | |||||
|
PDE4D_HUMAN
|
||||||
| θ value | 2.67181e-44 (rank : 4) | NC score | 0.922355 (rank : 13) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q08499, O43433, Q13549, Q13550, Q13551, Q7Z2L8, Q8IV84, Q8IVA9, Q8IVD2, Q8IVD3, Q9HCX7 | Gene names | PDE4D | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3) (PDE43). | |||||
|
PDE4D_MOUSE
|
||||||
| θ value | 2.67181e-44 (rank : 5) | NC score | 0.923069 (rank : 10) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q01063, Q6TRH9, Q8C4Q7, Q8CG05 | Gene names | Pde4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3). | |||||
|
PDE4A_HUMAN
|
||||||
| θ value | 7.77379e-44 (rank : 6) | NC score | 0.922698 (rank : 12) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27815, O75522, O76092, Q16255, Q16691, Q8WUQ3 | Gene names | PDE4A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17) (DPDE2) (PDE46). | |||||
|
PDE4A_MOUSE
|
||||||
| θ value | 1.73182e-43 (rank : 7) | NC score | 0.922923 (rank : 11) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O89084, Q8R078, Q9JHQ4, Q9QX48, Q9QX49, Q9QXI8 | Gene names | Pde4a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17). | |||||
|
PDE4C_HUMAN
|
||||||
| θ value | 1.91475e-42 (rank : 8) | NC score | 0.925802 (rank : 8) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08493, Q9UN44, Q9UN45, Q9UN46, Q9UPJ6 | Gene names | PDE4C | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17) (DPDE1) (PDE21). | |||||
|
PDE8A_HUMAN
|
||||||
| θ value | 9.50281e-42 (rank : 9) | NC score | 0.919362 (rank : 15) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O60658, Q969I1, Q96PC9, Q96PD0, Q96PD1, Q9UMB7 | Gene names | PDE8A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17). | |||||
|
PDE4C_MOUSE
|
||||||
| θ value | 1.62093e-41 (rank : 10) | NC score | 0.927002 (rank : 7) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3UEI1, Q8K0P4 | Gene names | Pde4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17). | |||||
|
PDE11_MOUSE
|
||||||
| θ value | 1.05066e-40 (rank : 11) | NC score | 0.890492 (rank : 26) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P0C1Q2 | Gene names | Pde11a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE11_HUMAN
|
||||||
| θ value | 2.34062e-40 (rank : 12) | NC score | 0.890528 (rank : 25) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HCR9, Q53T16, Q96S76, Q9GZY7, Q9HB46, Q9NY45 | Gene names | PDE11A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE8A_MOUSE
|
||||||
| θ value | 5.21438e-40 (rank : 13) | NC score | 0.915799 (rank : 20) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O88502 | Gene names | Pde8a, Pde8 | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17) (MMPDE8). | |||||
|
PDE7A_HUMAN
|
||||||
| θ value | 4.41421e-39 (rank : 14) | NC score | 0.935508 (rank : 3) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13946, O15380 | Gene names | PDE7A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (HCP1) (TM22). | |||||
|
PDE8B_HUMAN
|
||||||
| θ value | 4.41421e-39 (rank : 15) | NC score | 0.915944 (rank : 19) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95263, Q86XK8, Q8IUJ7, Q8IUJ8, Q8IUJ9, Q8IUK0, Q8N3T2 | Gene names | PDE8B | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8B (EC 3.1.4.17) (HSPDE8B). | |||||
|
PDE2A_HUMAN
|
||||||
| θ value | 1.6774e-38 (rank : 16) | NC score | 0.901562 (rank : 24) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00408 | Gene names | PDE2A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE2A_MOUSE
|
||||||
| θ value | 1.6774e-38 (rank : 17) | NC score | 0.902378 (rank : 23) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q922S4, Q8K2U1 | Gene names | Pde2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE7B_HUMAN
|
||||||
| θ value | 1.6774e-38 (rank : 18) | NC score | 0.932895 (rank : 6) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NP56 | Gene names | PDE7B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE7A_MOUSE
|
||||||
| θ value | 3.73686e-38 (rank : 19) | NC score | 0.934719 (rank : 4) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70453, Q9ERB3 | Gene names | Pde7a | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (P2A). | |||||
|
PDE7B_MOUSE
|
||||||
| θ value | 6.37413e-38 (rank : 20) | NC score | 0.933684 (rank : 5) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QXQ1 | Gene names | Pde7b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE1B_MOUSE
|
||||||
| θ value | 4.13158e-37 (rank : 21) | NC score | 0.920646 (rank : 14) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q01065, O35384 | Gene names | Pde1b, Pde1b1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE1A_MOUSE
|
||||||
| θ value | 7.04741e-37 (rank : 22) | NC score | 0.918526 (rank : 16) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61481, O35388 | Gene names | Pde1a | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE). | |||||
|
PDE1B_HUMAN
|
||||||
| θ value | 3.4976e-36 (rank : 23) | NC score | 0.918136 (rank : 17) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q01064, Q92825 | Gene names | PDE1B, PDE1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE1A_HUMAN
|
||||||
| θ value | 5.96599e-36 (rank : 24) | NC score | 0.916790 (rank : 18) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P54750, Q86VZ0, Q9C0K8, Q9C0K9, Q9C0L0, Q9C0L1, Q9C0L2, Q9C0L3, Q9C0L4, Q9UFX3 | Gene names | PDE1A | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE) (hCam-1). | |||||
|
PDE1C_HUMAN
|
||||||
| θ value | 2.26708e-35 (rank : 25) | NC score | 0.907003 (rank : 21) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
PDE1C_MOUSE
|
||||||
| θ value | 4.27553e-34 (rank : 26) | NC score | 0.905784 (rank : 22) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
PDE5A_HUMAN
|
||||||
| θ value | 1.52067e-31 (rank : 27) | NC score | 0.869507 (rank : 32) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O76074, O75026, O75887, Q86UI0, Q86V66, Q9Y6Z6 | Gene names | PDE5A, PDE5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE5A_MOUSE
|
||||||
| θ value | 1.52067e-31 (rank : 28) | NC score | 0.871677 (rank : 31) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CG03 | Gene names | Pde5a, Pde5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE6B_MOUSE
|
||||||
| θ value | 1.85889e-29 (rank : 29) | NC score | 0.845524 (rank : 37) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P23440 | Gene names | Pde6b, Mpb, Pdeb, rd | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PDE6B_HUMAN
|
||||||
| θ value | 5.40856e-29 (rank : 30) | NC score | 0.846765 (rank : 36) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35913, Q9BWH5 | Gene names | PDE6B, PDEB | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase beta-subunit precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PDE6A_HUMAN
|
||||||
| θ value | 1.57365e-28 (rank : 31) | NC score | 0.848350 (rank : 35) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16499 | Gene names | PDE6A, PDEA | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha) (PDE V-B1). | |||||
|
PDE3A_HUMAN
|
||||||
| θ value | 2.05525e-28 (rank : 32) | NC score | 0.878117 (rank : 29) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14432, O60865, Q13348 | Gene names | PDE3A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PDE3B_HUMAN
|
||||||
| θ value | 4.57862e-28 (rank : 33) | NC score | 0.888910 (rank : 27) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13370, O00639, Q14408 | Gene names | PDE3B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase B (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase B) (CGI-PDE B) (CGIPDE1) (CGIP1). | |||||
|
PDE3A_MOUSE
|
||||||
| θ value | 7.80994e-28 (rank : 34) | NC score | 0.876977 (rank : 30) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Z0X4 | Gene names | Pde3a | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PDE6C_MOUSE
|
||||||
| θ value | 7.80994e-28 (rank : 35) | NC score | 0.852280 (rank : 33) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91ZQ1, Q8R0D4 | Gene names | Pde6c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE6C_HUMAN
|
||||||
| θ value | 1.02001e-27 (rank : 36) | NC score | 0.851321 (rank : 34) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P51160, Q5VY29 | Gene names | PDE6C, PDEA2 | |||
|
Domain Architecture |

|
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE6A_MOUSE
|
||||||
| θ value | 3.28125e-26 (rank : 37) | NC score | 0.843867 (rank : 38) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27664 | Gene names | Pde6a, Mpa, Pdea | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha). | |||||
|
PDE10_HUMAN
|
||||||
| θ value | 5.59698e-26 (rank : 38) | NC score | 0.880106 (rank : 28) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y233, Q9Y5T1 | Gene names | PDE10A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A (EC 3.1.4.17). | |||||
|
SESN1_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 39) | NC score | 0.057115 (rank : 39) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58006, Q7TNF3 | Gene names | Sesn1, Pa26, Sest1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sestrin-1 (p53-regulated protein PA26). | |||||
|
PARC_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 40) | NC score | 0.040607 (rank : 41) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
SESN1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 41) | NC score | 0.048988 (rank : 40) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6P5, Q9NV00, Q9UPD5, Q9Y6P6 | Gene names | SESN1, PA26, SEST1 | |||
|
Domain Architecture |
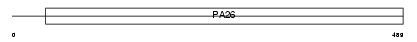
|
|||||
| Description | Sestrin-1 (p53-regulated protein PA26). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 42) | NC score | 0.022669 (rank : 44) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 0.21417 (rank : 43) | NC score | 0.022061 (rank : 49) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
PARC_MOUSE
|
||||||
| θ value | 0.279714 (rank : 44) | NC score | 0.034391 (rank : 42) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TT8, Q8BKL9, Q8CGC0, Q8R363 | Gene names | Parc, Kiaa0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein. | |||||
|
SMC4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 45) | NC score | 0.020964 (rank : 53) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
CCD41_MOUSE
|
||||||
| θ value | 0.47712 (rank : 46) | NC score | 0.015436 (rank : 61) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 47) | NC score | 0.022189 (rank : 47) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.022550 (rank : 45) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
BRE1B_HUMAN
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.021186 (rank : 51) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
BRE1B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.022173 (rank : 48) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.015607 (rank : 60) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.018228 (rank : 57) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
PSIP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.021106 (rank : 52) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99JF8, Q3TEJ7, Q3TTD7, Q3UTA1, Q80WQ7, Q99JF7, Q99LR4, Q9CT03 | Gene names | Psip1, Ledgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (mLEDGF). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.020667 (rank : 54) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
PSIP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.019112 (rank : 56) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75475, O00256, O95368, Q6P391, Q86YB9, Q9NZI3, Q9UER6 | Gene names | PSIP1, DFS70, LEDGF, PSIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (Transcriptional coactivator p75/p52) (Dense fine speckles 70 kDa protein) (DFS 70) (CLL-associated antigen KW-7). | |||||
|
K1C10_MOUSE
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.007243 (rank : 69) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P02535, P08731, Q8BUX3, Q8BV09, Q8BVU3, Q9CXH6, Q9JKB4 | Gene names | Krt10, Krt1-10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10) (56 kDa cytokeratin) (Keratin, type I cytoskeletal 59 kDa). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.022355 (rank : 46) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
TMF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.018108 (rank : 58) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.011507 (rank : 66) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.021660 (rank : 50) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
TRFL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.008293 (rank : 68) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08071, P70690, Q61799, Q922P2 | Gene names | Ltf | |||
|
Domain Architecture |

|
|||||
| Description | Lactotransferrin precursor (EC 3.4.21.-) (Lactoferrin). | |||||
|
CP2C8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.000774 (rank : 72) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P10632 | Gene names | CYP2C8 | |||
|
Domain Architecture |
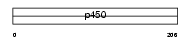
|
|||||
| Description | Cytochrome P450 2C8 (EC 1.14.14.1) (CYPIIC8) (P450 form 1) (P450 MP- 12/MP-20) (P450 IIC2) (S-mephenytoin 4-hydroxylase). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.013111 (rank : 63) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
K1C10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.006380 (rank : 71) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P13645, Q14664, Q8N175 | Gene names | KRT10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10). | |||||
|
PDCD7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.033928 (rank : 43) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WTY1, Q8R5D9 | Gene names | Pdcd7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death protein 7 (ES18). | |||||
|
SM1L2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.014342 (rank : 62) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
BICD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.012272 (rank : 65) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TD16, O75181, Q5TBQ2, Q5TBQ3, Q96LH2, Q9BT84, Q9H561 | Gene names | BICD2, KIAA0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
BICD2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.012810 (rank : 64) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.017077 (rank : 59) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.020407 (rank : 55) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
PIBF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.010767 (rank : 67) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
SAMH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.006787 (rank : 70) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y3Z3, Q5JXG8, Q9H004, Q9H005, Q9H3U9 | Gene names | SAMHD1, MOP5 | |||
|
Domain Architecture |
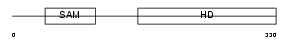
|
|||||
| Description | SAM domain and HD domain-containing protein 1 (Dendritic cell-derived IFNG-induced protein) (DCIP) (Monocyte protein 5) (MOP-5). | |||||
|
PDE9A_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O70628 | Gene names | Pde9a, Pde8b | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE9A_HUMAN
|
||||||
| NC score | 0.997194 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O76083, O75490, O75491, O95225, Q86SF7, Q86SI6, Q86SJ3, Q86WN3, Q86WN4, Q86WN5, Q86WN6, Q86WN7, Q86WN8, Q86WN9, Q86WP0 | Gene names | PDE9A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE7A_HUMAN
|
||||||
| NC score | 0.935508 (rank : 3) | θ value | 4.41421e-39 (rank : 14) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13946, O15380 | Gene names | PDE7A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (HCP1) (TM22). | |||||
|
PDE7A_MOUSE
|
||||||
| NC score | 0.934719 (rank : 4) | θ value | 3.73686e-38 (rank : 19) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70453, Q9ERB3 | Gene names | Pde7a | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (P2A). | |||||
|
PDE7B_MOUSE
|
||||||
| NC score | 0.933684 (rank : 5) | θ value | 6.37413e-38 (rank : 20) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QXQ1 | Gene names | Pde7b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE7B_HUMAN
|
||||||
| NC score | 0.932895 (rank : 6) | θ value | 1.6774e-38 (rank : 18) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NP56 | Gene names | PDE7B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE4C_MOUSE
|
||||||
| NC score | 0.927002 (rank : 7) | θ value | 1.62093e-41 (rank : 10) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3UEI1, Q8K0P4 | Gene names | Pde4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17). | |||||
|
PDE4C_HUMAN
|
||||||
| NC score | 0.925802 (rank : 8) | θ value | 1.91475e-42 (rank : 8) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08493, Q9UN44, Q9UN45, Q9UN46, Q9UPJ6 | Gene names | PDE4C | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17) (DPDE1) (PDE21). | |||||
|
PDE4B_HUMAN
|
||||||
| NC score | 0.924204 (rank : 9) | θ value | 1.41672e-45 (rank : 3) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q07343, O15443 | Gene names | PDE4B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4B (EC 3.1.4.17) (DPDE4) (PDE32). | |||||
|
PDE4D_MOUSE
|
||||||
| NC score | 0.923069 (rank : 10) | θ value | 2.67181e-44 (rank : 5) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q01063, Q6TRH9, Q8C4Q7, Q8CG05 | Gene names | Pde4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3). | |||||
|
PDE4A_MOUSE
|
||||||
| NC score | 0.922923 (rank : 11) | θ value | 1.73182e-43 (rank : 7) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O89084, Q8R078, Q9JHQ4, Q9QX48, Q9QX49, Q9QXI8 | Gene names | Pde4a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17). | |||||
|
PDE4A_HUMAN
|
||||||
| NC score | 0.922698 (rank : 12) | θ value | 7.77379e-44 (rank : 6) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27815, O75522, O76092, Q16255, Q16691, Q8WUQ3 | Gene names | PDE4A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17) (DPDE2) (PDE46). | |||||
|
PDE4D_HUMAN
|
||||||
| NC score | 0.922355 (rank : 13) | θ value | 2.67181e-44 (rank : 4) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q08499, O43433, Q13549, Q13550, Q13551, Q7Z2L8, Q8IV84, Q8IVA9, Q8IVD2, Q8IVD3, Q9HCX7 | Gene names | PDE4D | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3) (PDE43). | |||||
|
PDE1B_MOUSE
|
||||||
| NC score | 0.920646 (rank : 14) | θ value | 4.13158e-37 (rank : 21) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q01065, O35384 | Gene names | Pde1b, Pde1b1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE8A_HUMAN
|
||||||
| NC score | 0.919362 (rank : 15) | θ value | 9.50281e-42 (rank : 9) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O60658, Q969I1, Q96PC9, Q96PD0, Q96PD1, Q9UMB7 | Gene names | PDE8A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17). | |||||
|
PDE1A_MOUSE
|
||||||
| NC score | 0.918526 (rank : 16) | θ value | 7.04741e-37 (rank : 22) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61481, O35388 | Gene names | Pde1a | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE). | |||||
|
PDE1B_HUMAN
|
||||||
| NC score | 0.918136 (rank : 17) | θ value | 3.4976e-36 (rank : 23) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q01064, Q92825 | Gene names | PDE1B, PDE1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE1A_HUMAN
|
||||||
| NC score | 0.916790 (rank : 18) | θ value | 5.96599e-36 (rank : 24) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P54750, Q86VZ0, Q9C0K8, Q9C0K9, Q9C0L0, Q9C0L1, Q9C0L2, Q9C0L3, Q9C0L4, Q9UFX3 | Gene names | PDE1A | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE) (hCam-1). | |||||
|
PDE8B_HUMAN
|
||||||
| NC score | 0.915944 (rank : 19) | θ value | 4.41421e-39 (rank : 15) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95263, Q86XK8, Q8IUJ7, Q8IUJ8, Q8IUJ9, Q8IUK0, Q8N3T2 | Gene names | PDE8B | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8B (EC 3.1.4.17) (HSPDE8B). | |||||
|
PDE8A_MOUSE
|
||||||
| NC score | 0.915799 (rank : 20) | θ value | 5.21438e-40 (rank : 13) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O88502 | Gene names | Pde8a, Pde8 | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17) (MMPDE8). | |||||
|
PDE1C_HUMAN
|
||||||
| NC score | 0.907003 (rank : 21) | θ value | 2.26708e-35 (rank : 25) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
PDE1C_MOUSE
|
||||||
| NC score | 0.905784 (rank : 22) | θ value | 4.27553e-34 (rank : 26) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
PDE2A_MOUSE
|
||||||
| NC score | 0.902378 (rank : 23) | θ value | 1.6774e-38 (rank : 17) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q922S4, Q8K2U1 | Gene names | Pde2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE2A_HUMAN
|
||||||
| NC score | 0.901562 (rank : 24) | θ value | 1.6774e-38 (rank : 16) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00408 | Gene names | PDE2A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE11_HUMAN
|
||||||
| NC score | 0.890528 (rank : 25) | θ value | 2.34062e-40 (rank : 12) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HCR9, Q53T16, Q96S76, Q9GZY7, Q9HB46, Q9NY45 | Gene names | PDE11A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE11_MOUSE
|
||||||
| NC score | 0.890492 (rank : 26) | θ value | 1.05066e-40 (rank : 11) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P0C1Q2 | Gene names | Pde11a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE3B_HUMAN
|
||||||
| NC score | 0.888910 (rank : 27) | θ value | 4.57862e-28 (rank : 33) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13370, O00639, Q14408 | Gene names | PDE3B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase B (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase B) (CGI-PDE B) (CGIPDE1) (CGIP1). | |||||
|
PDE10_HUMAN
|
||||||
| NC score | 0.880106 (rank : 28) | θ value | 5.59698e-26 (rank : 38) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y233, Q9Y5T1 | Gene names | PDE10A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A (EC 3.1.4.17). | |||||
|
PDE3A_HUMAN
|
||||||
| NC score | 0.878117 (rank : 29) | θ value | 2.05525e-28 (rank : 32) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14432, O60865, Q13348 | Gene names | PDE3A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PDE3A_MOUSE
|
||||||
| NC score | 0.876977 (rank : 30) | θ value | 7.80994e-28 (rank : 34) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Z0X4 | Gene names | Pde3a | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PDE5A_MOUSE
|
||||||
| NC score | 0.871677 (rank : 31) | θ value | 1.52067e-31 (rank : 28) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CG03 | Gene names | Pde5a, Pde5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE5A_HUMAN
|
||||||
| NC score | 0.869507 (rank : 32) | θ value | 1.52067e-31 (rank : 27) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O76074, O75026, O75887, Q86UI0, Q86V66, Q9Y6Z6 | Gene names | PDE5A, PDE5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE6C_MOUSE
|
||||||
| NC score | 0.852280 (rank : 33) | θ value | 7.80994e-28 (rank : 35) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91ZQ1, Q8R0D4 | Gene names | Pde6c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE6C_HUMAN
|
||||||
| NC score | 0.851321 (rank : 34) | θ value | 1.02001e-27 (rank : 36) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P51160, Q5VY29 | Gene names | PDE6C, PDEA2 | |||
|
Domain Architecture |

|
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE6A_HUMAN
|
||||||
| NC score | 0.848350 (rank : 35) | θ value | 1.57365e-28 (rank : 31) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16499 | Gene names | PDE6A, PDEA | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha) (PDE V-B1). | |||||
|
PDE6B_HUMAN
|
||||||
| NC score | 0.846765 (rank : 36) | θ value | 5.40856e-29 (rank : 30) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35913, Q9BWH5 | Gene names | PDE6B, PDEB | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase beta-subunit precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PDE6B_MOUSE
|
||||||
| NC score | 0.845524 (rank : 37) | θ value | 1.85889e-29 (rank : 29) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P23440 | Gene names | Pde6b, Mpb, Pdeb, rd | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PDE6A_MOUSE
|
||||||
| NC score | 0.843867 (rank : 38) | θ value | 3.28125e-26 (rank : 37) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27664 | Gene names | Pde6a, Mpa, Pdea | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha). | |||||
|
SESN1_MOUSE
|
||||||
| NC score | 0.057115 (rank : 39) | θ value | 0.00390308 (rank : 39) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58006, Q7TNF3 | Gene names | Sesn1, Pa26, Sest1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sestrin-1 (p53-regulated protein PA26). | |||||
|
SESN1_HUMAN
|
||||||
| NC score | 0.048988 (rank : 40) | θ value | 0.0736092 (rank : 41) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6P5, Q9NV00, Q9UPD5, Q9Y6P6 | Gene names | SESN1, PA26, SEST1 | |||
|
Domain Architecture |

|
|||||
| Description | Sestrin-1 (p53-regulated protein PA26). | |||||
|
PARC_HUMAN
|
||||||
| NC score | 0.040607 (rank : 41) | θ value | 0.0252991 (rank : 40) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
PARC_MOUSE
|
||||||
| NC score | 0.034391 (rank : 42) | θ value | 0.279714 (rank : 44) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TT8, Q8BKL9, Q8CGC0, Q8R363 | Gene names | Parc, Kiaa0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein. | |||||
|
PDCD7_MOUSE
|
||||||
| NC score | 0.033928 (rank : 43) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WTY1, Q8R5D9 | Gene names | Pdcd7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death protein 7 (ES18). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.022669 (rank : 44) | θ value | 0.21417 (rank : 42) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.022550 (rank : 45) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.022355 (rank : 46) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.022189 (rank : 47) | θ value | 0.47712 (rank : 47) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
BRE1B_MOUSE
|
||||||
| NC score | 0.022173 (rank : 48) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.022061 (rank : 49) | θ value | 0.21417 (rank : 43) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.021660 (rank : 50) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
BRE1B_HUMAN
|
||||||
| NC score | 0.021186 (rank : 51) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
PSIP1_MOUSE
|
||||||
| NC score | 0.021106 (rank : 52) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99JF8, Q3TEJ7, Q3TTD7, Q3UTA1, Q80WQ7, Q99JF7, Q99LR4, Q9CT03 | Gene names | Psip1, Ledgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (mLEDGF). | |||||
|
SMC4_HUMAN
|
||||||
| NC score | 0.020964 (rank : 53) | θ value | 0.365318 (rank : 45) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.020667 (rank : 54) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.020407 (rank : 55) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
PSIP1_HUMAN
|
||||||
| NC score | 0.019112 (rank : 56) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75475, O00256, O95368, Q6P391, Q86YB9, Q9NZI3, Q9UER6 | Gene names | PSIP1, DFS70, LEDGF, PSIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (Transcriptional coactivator p75/p52) (Dense fine speckles 70 kDa protein) (DFS 70) (CLL-associated antigen KW-7). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.018228 (rank : 57) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
TMF1_HUMAN
|
||||||
| NC score | 0.018108 (rank : 58) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.017077 (rank : 59) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.015607 (rank : 60) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.015436 (rank : 61) | θ value | 0.47712 (rank : 46) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
SM1L2_HUMAN
|
||||||
| NC score | 0.014342 (rank : 62) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.013111 (rank : 63) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
BICD2_MOUSE
|
||||||
| NC score | 0.012810 (rank : 64) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
BICD2_HUMAN
|
||||||
| NC score | 0.012272 (rank : 65) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TD16, O75181, Q5TBQ2, Q5TBQ3, Q96LH2, Q9BT84, Q9H561 | Gene names | BICD2, KIAA0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.011507 (rank : 66) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
PIBF1_HUMAN
|
||||||
| NC score | 0.010767 (rank : 67) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
TRFL_MOUSE
|
||||||
| NC score | 0.008293 (rank : 68) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08071, P70690, Q61799, Q922P2 | Gene names | Ltf | |||
|
Domain Architecture |

|
|||||
| Description | Lactotransferrin precursor (EC 3.4.21.-) (Lactoferrin). | |||||
|
K1C10_MOUSE
|
||||||
| NC score | 0.007243 (rank : 69) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P02535, P08731, Q8BUX3, Q8BV09, Q8BVU3, Q9CXH6, Q9JKB4 | Gene names | Krt10, Krt1-10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10) (56 kDa cytokeratin) (Keratin, type I cytoskeletal 59 kDa). | |||||
|
SAMH1_HUMAN
|
||||||
| NC score | 0.006787 (rank : 70) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y3Z3, Q5JXG8, Q9H004, Q9H005, Q9H3U9 | Gene names | SAMHD1, MOP5 | |||
|
Domain Architecture |

|
|||||
| Description | SAM domain and HD domain-containing protein 1 (Dendritic cell-derived IFNG-induced protein) (DCIP) (Monocyte protein 5) (MOP-5). | |||||
|
K1C10_HUMAN
|
||||||
| NC score | 0.006380 (rank : 71) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P13645, Q14664, Q8N175 | Gene names | KRT10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10). | |||||
|
CP2C8_HUMAN
|
||||||
| NC score | 0.000774 (rank : 72) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P10632 | Gene names | CYP2C8 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2C8 (EC 1.14.14.1) (CYPIIC8) (P450 form 1) (P450 MP- 12/MP-20) (P450 IIC2) (S-mephenytoin 4-hydroxylase). | |||||