Please be patient as the page loads
|
PDE11_MOUSE
|
||||||
| SwissProt Accessions | P0C1Q2 | Gene names | Pde11a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PDE11_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999618 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9HCR9, Q53T16, Q96S76, Q9GZY7, Q9HB46, Q9NY45 | Gene names | PDE11A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE11_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P0C1Q2 | Gene names | Pde11a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE5A_HUMAN
|
||||||
| θ value | 9.78303e-172 (rank : 3) | NC score | 0.983904 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O76074, O75026, O75887, Q86UI0, Q86V66, Q9Y6Z6 | Gene names | PDE5A, PDE5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE5A_MOUSE
|
||||||
| θ value | 4.54438e-169 (rank : 4) | NC score | 0.984365 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CG03 | Gene names | Pde5a, Pde5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE6A_HUMAN
|
||||||
| θ value | 1.21265e-121 (rank : 5) | NC score | 0.962683 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16499 | Gene names | PDE6A, PDEA | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha) (PDE V-B1). | |||||
|
PDE6B_HUMAN
|
||||||
| θ value | 2.06847e-121 (rank : 6) | NC score | 0.963540 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35913, Q9BWH5 | Gene names | PDE6B, PDEB | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase beta-subunit precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PDE6A_MOUSE
|
||||||
| θ value | 1.75107e-120 (rank : 7) | NC score | 0.961791 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27664 | Gene names | Pde6a, Mpa, Pdea | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha). | |||||
|
PDE6B_MOUSE
|
||||||
| θ value | 1.75107e-120 (rank : 8) | NC score | 0.962525 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P23440 | Gene names | Pde6b, Mpb, Pdeb, rd | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PDE6C_HUMAN
|
||||||
| θ value | 9.94316e-116 (rank : 9) | NC score | 0.963381 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P51160, Q5VY29 | Gene names | PDE6C, PDEA2 | |||
|
Domain Architecture |

|
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE6C_MOUSE
|
||||||
| θ value | 9.94316e-116 (rank : 10) | NC score | 0.964473 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91ZQ1, Q8R0D4 | Gene names | Pde6c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE10_HUMAN
|
||||||
| θ value | 1.3939e-101 (rank : 11) | NC score | 0.975336 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y233, Q9Y5T1 | Gene names | PDE10A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A (EC 3.1.4.17). | |||||
|
PDE2A_MOUSE
|
||||||
| θ value | 6.47494e-99 (rank : 12) | NC score | 0.962390 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q922S4, Q8K2U1 | Gene names | Pde2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE2A_HUMAN
|
||||||
| θ value | 1.10445e-98 (rank : 13) | NC score | 0.961313 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00408 | Gene names | PDE2A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE9A_MOUSE
|
||||||
| θ value | 1.05066e-40 (rank : 14) | NC score | 0.890492 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O70628 | Gene names | Pde9a, Pde8b | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE9A_HUMAN
|
||||||
| θ value | 8.89434e-40 (rank : 15) | NC score | 0.889898 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O76083, O75490, O75491, O95225, Q86SF7, Q86SI6, Q86SJ3, Q86WN3, Q86WN4, Q86WN5, Q86WN6, Q86WN7, Q86WN8, Q86WN9, Q86WP0 | Gene names | PDE9A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE4C_MOUSE
|
||||||
| θ value | 4.27553e-34 (rank : 16) | NC score | 0.838883 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3UEI1, Q8K0P4 | Gene names | Pde4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17). | |||||
|
PDE4D_HUMAN
|
||||||
| θ value | 1.0531e-32 (rank : 17) | NC score | 0.831248 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08499, O43433, Q13549, Q13550, Q13551, Q7Z2L8, Q8IV84, Q8IVA9, Q8IVD2, Q8IVD3, Q9HCX7 | Gene names | PDE4D | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3) (PDE43). | |||||
|
PDE4D_MOUSE
|
||||||
| θ value | 1.79631e-32 (rank : 18) | NC score | 0.831927 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01063, Q6TRH9, Q8C4Q7, Q8CG05 | Gene names | Pde4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3). | |||||
|
PDE4A_HUMAN
|
||||||
| θ value | 3.06405e-32 (rank : 19) | NC score | 0.830845 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P27815, O75522, O76092, Q16255, Q16691, Q8WUQ3 | Gene names | PDE4A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17) (DPDE2) (PDE46). | |||||
|
PDE4A_MOUSE
|
||||||
| θ value | 3.06405e-32 (rank : 20) | NC score | 0.831239 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O89084, Q8R078, Q9JHQ4, Q9QX48, Q9QX49, Q9QXI8 | Gene names | Pde4a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17). | |||||
|
PDE1B_MOUSE
|
||||||
| θ value | 5.22648e-32 (rank : 21) | NC score | 0.832165 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01065, O35384 | Gene names | Pde1b, Pde1b1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE4C_HUMAN
|
||||||
| θ value | 5.22648e-32 (rank : 22) | NC score | 0.836978 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08493, Q9UN44, Q9UN45, Q9UN46, Q9UPJ6 | Gene names | PDE4C | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17) (DPDE1) (PDE21). | |||||
|
PDE7A_HUMAN
|
||||||
| θ value | 1.98606e-31 (rank : 23) | NC score | 0.853568 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13946, O15380 | Gene names | PDE7A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (HCP1) (TM22). | |||||
|
PDE7A_MOUSE
|
||||||
| θ value | 1.98606e-31 (rank : 24) | NC score | 0.851399 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70453, Q9ERB3 | Gene names | Pde7a | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (P2A). | |||||
|
PDE1B_HUMAN
|
||||||
| θ value | 3.3877e-31 (rank : 25) | NC score | 0.828684 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q01064, Q92825 | Gene names | PDE1B, PDE1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE7B_MOUSE
|
||||||
| θ value | 3.3877e-31 (rank : 26) | NC score | 0.852868 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QXQ1 | Gene names | Pde7b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE4B_HUMAN
|
||||||
| θ value | 7.54701e-31 (rank : 27) | NC score | 0.826382 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q07343, O15443 | Gene names | PDE4B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4B (EC 3.1.4.17) (DPDE4) (PDE32). | |||||
|
PDE7B_HUMAN
|
||||||
| θ value | 3.74554e-30 (rank : 28) | NC score | 0.850532 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NP56 | Gene names | PDE7B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE1A_MOUSE
|
||||||
| θ value | 2.27234e-27 (rank : 29) | NC score | 0.828110 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61481, O35388 | Gene names | Pde1a | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE). | |||||
|
PDE1C_HUMAN
|
||||||
| θ value | 2.96777e-27 (rank : 30) | NC score | 0.817847 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
PDE1C_MOUSE
|
||||||
| θ value | 2.96777e-27 (rank : 31) | NC score | 0.816990 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
PDE1A_HUMAN
|
||||||
| θ value | 5.06226e-27 (rank : 32) | NC score | 0.825731 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P54750, Q86VZ0, Q9C0K8, Q9C0K9, Q9C0L0, Q9C0L1, Q9C0L2, Q9C0L3, Q9C0L4, Q9UFX3 | Gene names | PDE1A | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE) (hCam-1). | |||||
|
PDE8A_MOUSE
|
||||||
| θ value | 2.51237e-26 (rank : 33) | NC score | 0.809650 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88502 | Gene names | Pde8a, Pde8 | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17) (MMPDE8). | |||||
|
PDE8A_HUMAN
|
||||||
| θ value | 5.59698e-26 (rank : 34) | NC score | 0.814217 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O60658, Q969I1, Q96PC9, Q96PD0, Q96PD1, Q9UMB7 | Gene names | PDE8A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17). | |||||
|
PDE8B_HUMAN
|
||||||
| θ value | 4.01107e-24 (rank : 35) | NC score | 0.806912 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95263, Q86XK8, Q8IUJ7, Q8IUJ8, Q8IUJ9, Q8IUK0, Q8N3T2 | Gene names | PDE8B | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8B (EC 3.1.4.17) (HSPDE8B). | |||||
|
PDE3A_MOUSE
|
||||||
| θ value | 1.13037e-18 (rank : 36) | NC score | 0.767562 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Z0X4 | Gene names | Pde3a | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PDE3A_HUMAN
|
||||||
| θ value | 1.92812e-18 (rank : 37) | NC score | 0.764956 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14432, O60865, Q13348 | Gene names | PDE3A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PDE3B_HUMAN
|
||||||
| θ value | 3.28887e-18 (rank : 38) | NC score | 0.773481 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13370, O00639, Q14408 | Gene names | PDE3B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase B (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase B) (CGI-PDE B) (CGIPDE1) (CGIP1). | |||||
|
CO4A2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.003479 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
PMF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.019104 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CPV5, Q3UCB8, Q8K237, Q924B3, Q9D6E2 | Gene names | Pmf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyamine-modulated factor 1 (PMF-1). | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.000468 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
CO9A3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.003398 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
JHD2B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.006320 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZPY7, Q2VPQ5, Q5U5V7, Q6P9K3, Q8CCE2, Q8K2A5, Q9CU57 | Gene names | Jmjd1b, Jhdm2b, Kiaa1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B). | |||||
|
EPHA5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | -0.003039 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54756 | Gene names | EPHA5, EHK1, HEK7 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 5 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EHK-1) (EPH homology kinase 1) (Receptor protein- tyrosine kinase HEK7). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | -0.001305 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
ZN431_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | -0.004127 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TF32, Q8IWC4 | Gene names | ZNF431, KIAA1969 | |||
|
Domain Architecture |
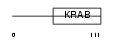
|
|||||
| Description | Zinc finger protein 431. | |||||
|
CO4A6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.002923 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14031, Q12823, Q14053, Q5JYH6, Q9NQM5, Q9NTX3, Q9UJ76, Q9UMG6, Q9Y4L4 | Gene names | COL4A6 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-6(IV) chain precursor. | |||||
|
SLK_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | -0.004603 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
SLK_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | -0.004789 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
BTBD7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.003797 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P203, Q69Z05, Q7Z308, Q86TS0, Q9HAA4, Q9NVM0 | Gene names | BTBD7, KIAA1525 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 7. | |||||
|
PDE11_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P0C1Q2 | Gene names | Pde11a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE11_HUMAN
|
||||||
| NC score | 0.999618 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9HCR9, Q53T16, Q96S76, Q9GZY7, Q9HB46, Q9NY45 | Gene names | PDE11A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE5A_MOUSE
|
||||||
| NC score | 0.984365 (rank : 3) | θ value | 4.54438e-169 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CG03 | Gene names | Pde5a, Pde5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE5A_HUMAN
|
||||||
| NC score | 0.983904 (rank : 4) | θ value | 9.78303e-172 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O76074, O75026, O75887, Q86UI0, Q86V66, Q9Y6Z6 | Gene names | PDE5A, PDE5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE10_HUMAN
|
||||||
| NC score | 0.975336 (rank : 5) | θ value | 1.3939e-101 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y233, Q9Y5T1 | Gene names | PDE10A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A (EC 3.1.4.17). | |||||
|
PDE6C_MOUSE
|
||||||
| NC score | 0.964473 (rank : 6) | θ value | 9.94316e-116 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91ZQ1, Q8R0D4 | Gene names | Pde6c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE6B_HUMAN
|
||||||
| NC score | 0.963540 (rank : 7) | θ value | 2.06847e-121 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35913, Q9BWH5 | Gene names | PDE6B, PDEB | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase beta-subunit precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PDE6C_HUMAN
|
||||||
| NC score | 0.963381 (rank : 8) | θ value | 9.94316e-116 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P51160, Q5VY29 | Gene names | PDE6C, PDEA2 | |||
|
Domain Architecture |

|
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE6A_HUMAN
|
||||||
| NC score | 0.962683 (rank : 9) | θ value | 1.21265e-121 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16499 | Gene names | PDE6A, PDEA | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha) (PDE V-B1). | |||||
|
PDE6B_MOUSE
|
||||||
| NC score | 0.962525 (rank : 10) | θ value | 1.75107e-120 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P23440 | Gene names | Pde6b, Mpb, Pdeb, rd | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PDE2A_MOUSE
|
||||||
| NC score | 0.962390 (rank : 11) | θ value | 6.47494e-99 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q922S4, Q8K2U1 | Gene names | Pde2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE6A_MOUSE
|
||||||
| NC score | 0.961791 (rank : 12) | θ value | 1.75107e-120 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27664 | Gene names | Pde6a, Mpa, Pdea | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha). | |||||
|
PDE2A_HUMAN
|
||||||
| NC score | 0.961313 (rank : 13) | θ value | 1.10445e-98 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00408 | Gene names | PDE2A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE9A_MOUSE
|
||||||
| NC score | 0.890492 (rank : 14) | θ value | 1.05066e-40 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O70628 | Gene names | Pde9a, Pde8b | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE9A_HUMAN
|
||||||
| NC score | 0.889898 (rank : 15) | θ value | 8.89434e-40 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O76083, O75490, O75491, O95225, Q86SF7, Q86SI6, Q86SJ3, Q86WN3, Q86WN4, Q86WN5, Q86WN6, Q86WN7, Q86WN8, Q86WN9, Q86WP0 | Gene names | PDE9A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE7A_HUMAN
|
||||||
| NC score | 0.853568 (rank : 16) | θ value | 1.98606e-31 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13946, O15380 | Gene names | PDE7A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (HCP1) (TM22). | |||||
|
PDE7B_MOUSE
|
||||||
| NC score | 0.852868 (rank : 17) | θ value | 3.3877e-31 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QXQ1 | Gene names | Pde7b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE7A_MOUSE
|
||||||
| NC score | 0.851399 (rank : 18) | θ value | 1.98606e-31 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70453, Q9ERB3 | Gene names | Pde7a | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (P2A). | |||||
|
PDE7B_HUMAN
|
||||||
| NC score | 0.850532 (rank : 19) | θ value | 3.74554e-30 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NP56 | Gene names | PDE7B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE4C_MOUSE
|
||||||
| NC score | 0.838883 (rank : 20) | θ value | 4.27553e-34 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3UEI1, Q8K0P4 | Gene names | Pde4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17). | |||||
|
PDE4C_HUMAN
|
||||||
| NC score | 0.836978 (rank : 21) | θ value | 5.22648e-32 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08493, Q9UN44, Q9UN45, Q9UN46, Q9UPJ6 | Gene names | PDE4C | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17) (DPDE1) (PDE21). | |||||
|
PDE1B_MOUSE
|
||||||
| NC score | 0.832165 (rank : 22) | θ value | 5.22648e-32 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01065, O35384 | Gene names | Pde1b, Pde1b1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE4D_MOUSE
|
||||||
| NC score | 0.831927 (rank : 23) | θ value | 1.79631e-32 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01063, Q6TRH9, Q8C4Q7, Q8CG05 | Gene names | Pde4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3). | |||||
|
PDE4D_HUMAN
|
||||||
| NC score | 0.831248 (rank : 24) | θ value | 1.0531e-32 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08499, O43433, Q13549, Q13550, Q13551, Q7Z2L8, Q8IV84, Q8IVA9, Q8IVD2, Q8IVD3, Q9HCX7 | Gene names | PDE4D | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3) (PDE43). | |||||
|
PDE4A_MOUSE
|
||||||
| NC score | 0.831239 (rank : 25) | θ value | 3.06405e-32 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O89084, Q8R078, Q9JHQ4, Q9QX48, Q9QX49, Q9QXI8 | Gene names | Pde4a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17). | |||||
|
PDE4A_HUMAN
|
||||||
| NC score | 0.830845 (rank : 26) | θ value | 3.06405e-32 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P27815, O75522, O76092, Q16255, Q16691, Q8WUQ3 | Gene names | PDE4A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17) (DPDE2) (PDE46). | |||||
|
PDE1B_HUMAN
|
||||||
| NC score | 0.828684 (rank : 27) | θ value | 3.3877e-31 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q01064, Q92825 | Gene names | PDE1B, PDE1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE1A_MOUSE
|
||||||
| NC score | 0.828110 (rank : 28) | θ value | 2.27234e-27 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61481, O35388 | Gene names | Pde1a | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE). | |||||
|
PDE4B_HUMAN
|
||||||
| NC score | 0.826382 (rank : 29) | θ value | 7.54701e-31 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q07343, O15443 | Gene names | PDE4B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4B (EC 3.1.4.17) (DPDE4) (PDE32). | |||||
|
PDE1A_HUMAN
|
||||||
| NC score | 0.825731 (rank : 30) | θ value | 5.06226e-27 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P54750, Q86VZ0, Q9C0K8, Q9C0K9, Q9C0L0, Q9C0L1, Q9C0L2, Q9C0L3, Q9C0L4, Q9UFX3 | Gene names | PDE1A | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE) (hCam-1). | |||||
|
PDE1C_HUMAN
|
||||||
| NC score | 0.817847 (rank : 31) | θ value | 2.96777e-27 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
PDE1C_MOUSE
|
||||||
| NC score | 0.816990 (rank : 32) | θ value | 2.96777e-27 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
PDE8A_HUMAN
|
||||||
| NC score | 0.814217 (rank : 33) | θ value | 5.59698e-26 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O60658, Q969I1, Q96PC9, Q96PD0, Q96PD1, Q9UMB7 | Gene names | PDE8A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17). | |||||
|
PDE8A_MOUSE
|
||||||
| NC score | 0.809650 (rank : 34) | θ value | 2.51237e-26 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88502 | Gene names | Pde8a, Pde8 | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17) (MMPDE8). | |||||
|
PDE8B_HUMAN
|
||||||
| NC score | 0.806912 (rank : 35) | θ value | 4.01107e-24 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95263, Q86XK8, Q8IUJ7, Q8IUJ8, Q8IUJ9, Q8IUK0, Q8N3T2 | Gene names | PDE8B | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8B (EC 3.1.4.17) (HSPDE8B). | |||||
|
PDE3B_HUMAN
|
||||||
| NC score | 0.773481 (rank : 36) | θ value | 3.28887e-18 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13370, O00639, Q14408 | Gene names | PDE3B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase B (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase B) (CGI-PDE B) (CGIPDE1) (CGIP1). | |||||
|
PDE3A_MOUSE
|
||||||
| NC score | 0.767562 (rank : 37) | θ value | 1.13037e-18 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Z0X4 | Gene names | Pde3a | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PDE3A_HUMAN
|
||||||
| NC score | 0.764956 (rank : 38) | θ value | 1.92812e-18 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14432, O60865, Q13348 | Gene names | PDE3A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PMF1_MOUSE
|
||||||
| NC score | 0.019104 (rank : 39) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CPV5, Q3UCB8, Q8K237, Q924B3, Q9D6E2 | Gene names | Pmf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyamine-modulated factor 1 (PMF-1). | |||||
|
JHD2B_MOUSE
|
||||||
| NC score | 0.006320 (rank : 40) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZPY7, Q2VPQ5, Q5U5V7, Q6P9K3, Q8CCE2, Q8K2A5, Q9CU57 | Gene names | Jmjd1b, Jhdm2b, Kiaa1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B). | |||||
|
BTBD7_HUMAN
|
||||||
| NC score | 0.003797 (rank : 41) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P203, Q69Z05, Q7Z308, Q86TS0, Q9HAA4, Q9NVM0 | Gene names | BTBD7, KIAA1525 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 7. | |||||
|
CO4A2_HUMAN
|
||||||
| NC score | 0.003479 (rank : 42) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
CO9A3_HUMAN
|
||||||
| NC score | 0.003398 (rank : 43) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
CO4A6_HUMAN
|
||||||
| NC score | 0.002923 (rank : 44) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14031, Q12823, Q14053, Q5JYH6, Q9NQM5, Q9NTX3, Q9UJ76, Q9UMG6, Q9Y4L4 | Gene names | COL4A6 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-6(IV) chain precursor. | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.000468 (rank : 45) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
MYH13_HUMAN
|
||||||
| NC score | -0.001305 (rank : 46) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
EPHA5_HUMAN
|
||||||
| NC score | -0.003039 (rank : 47) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54756 | Gene names | EPHA5, EHK1, HEK7 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 5 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EHK-1) (EPH homology kinase 1) (Receptor protein- tyrosine kinase HEK7). | |||||
|
ZN431_HUMAN
|
||||||
| NC score | -0.004127 (rank : 48) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TF32, Q8IWC4 | Gene names | ZNF431, KIAA1969 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 431. | |||||
|
SLK_HUMAN
|
||||||
| NC score | -0.004603 (rank : 49) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
SLK_MOUSE
|
||||||
| NC score | -0.004789 (rank : 50) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||