Please be patient as the page loads
|
PDE6C_HUMAN
|
||||||
| SwissProt Accessions | P51160, Q5VY29 | Gene names | PDE6C, PDEA2 | |||
|
Domain Architecture |

|
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PDE6A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.996767 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16499 | Gene names | PDE6A, PDEA | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha) (PDE V-B1). | |||||
|
PDE6A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.996882 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P27664 | Gene names | Pde6a, Mpa, Pdea | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha). | |||||
|
PDE6B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.997365 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35913, Q9BWH5 | Gene names | PDE6B, PDEB | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase beta-subunit precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PDE6B_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.997253 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P23440 | Gene names | Pde6b, Mpb, Pdeb, rd | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PDE6C_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P51160, Q5VY29 | Gene names | PDE6C, PDEA2 | |||
|
Domain Architecture |

|
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE6C_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.999359 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q91ZQ1, Q8R0D4 | Gene names | Pde6c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE11_MOUSE
|
||||||
| θ value | 9.94316e-116 (rank : 7) | NC score | 0.963381 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P0C1Q2 | Gene names | Pde11a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE11_HUMAN
|
||||||
| θ value | 1.29861e-115 (rank : 8) | NC score | 0.963448 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HCR9, Q53T16, Q96S76, Q9GZY7, Q9HB46, Q9NY45 | Gene names | PDE11A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE5A_MOUSE
|
||||||
| θ value | 8.73085e-104 (rank : 9) | NC score | 0.960155 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CG03 | Gene names | Pde5a, Pde5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE5A_HUMAN
|
||||||
| θ value | 1.26073e-102 (rank : 10) | NC score | 0.959139 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O76074, O75026, O75887, Q86UI0, Q86V66, Q9Y6Z6 | Gene names | PDE5A, PDE5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE2A_MOUSE
|
||||||
| θ value | 2.65329e-68 (rank : 11) | NC score | 0.933088 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q922S4, Q8K2U1 | Gene names | Pde2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE2A_HUMAN
|
||||||
| θ value | 3.4653e-68 (rank : 12) | NC score | 0.931672 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O00408 | Gene names | PDE2A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE10_HUMAN
|
||||||
| θ value | 1.27544e-62 (rank : 13) | NC score | 0.944663 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y233, Q9Y5T1 | Gene names | PDE10A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A (EC 3.1.4.17). | |||||
|
PDE4D_HUMAN
|
||||||
| θ value | 1.6247e-33 (rank : 14) | NC score | 0.806558 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08499, O43433, Q13549, Q13550, Q13551, Q7Z2L8, Q8IV84, Q8IVA9, Q8IVD2, Q8IVD3, Q9HCX7 | Gene names | PDE4D | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3) (PDE43). | |||||
|
PDE4A_HUMAN
|
||||||
| θ value | 2.12192e-33 (rank : 15) | NC score | 0.806304 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27815, O75522, O76092, Q16255, Q16691, Q8WUQ3 | Gene names | PDE4A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17) (DPDE2) (PDE46). | |||||
|
PDE4A_MOUSE
|
||||||
| θ value | 2.77131e-33 (rank : 16) | NC score | 0.806643 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O89084, Q8R078, Q9JHQ4, Q9QX48, Q9QX49, Q9QXI8 | Gene names | Pde4a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17). | |||||
|
PDE4D_MOUSE
|
||||||
| θ value | 2.77131e-33 (rank : 17) | NC score | 0.807305 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01063, Q6TRH9, Q8C4Q7, Q8CG05 | Gene names | Pde4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3). | |||||
|
PDE9A_HUMAN
|
||||||
| θ value | 1.08979e-29 (rank : 18) | NC score | 0.853302 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O76083, O75490, O75491, O95225, Q86SF7, Q86SI6, Q86SJ3, Q86WN3, Q86WN4, Q86WN5, Q86WN6, Q86WN7, Q86WN8, Q86WN9, Q86WP0 | Gene names | PDE9A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE4C_HUMAN
|
||||||
| θ value | 7.06379e-29 (rank : 19) | NC score | 0.809267 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08493, Q9UN44, Q9UN45, Q9UN46, Q9UPJ6 | Gene names | PDE4C | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17) (DPDE1) (PDE21). | |||||
|
PDE4B_HUMAN
|
||||||
| θ value | 1.57365e-28 (rank : 20) | NC score | 0.798275 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q07343, O15443 | Gene names | PDE4B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4B (EC 3.1.4.17) (DPDE4) (PDE32). | |||||
|
PDE4C_MOUSE
|
||||||
| θ value | 3.50572e-28 (rank : 21) | NC score | 0.809858 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3UEI1, Q8K0P4 | Gene names | Pde4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17). | |||||
|
PDE9A_MOUSE
|
||||||
| θ value | 1.02001e-27 (rank : 22) | NC score | 0.851321 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O70628 | Gene names | Pde9a, Pde8b | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE1A_MOUSE
|
||||||
| θ value | 8.63488e-27 (rank : 23) | NC score | 0.798865 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61481, O35388 | Gene names | Pde1a | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE). | |||||
|
PDE1A_HUMAN
|
||||||
| θ value | 2.51237e-26 (rank : 24) | NC score | 0.796528 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P54750, Q86VZ0, Q9C0K8, Q9C0K9, Q9C0L0, Q9C0L1, Q9C0L2, Q9C0L3, Q9C0L4, Q9UFX3 | Gene names | PDE1A | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE) (hCam-1). | |||||
|
PDE1C_HUMAN
|
||||||
| θ value | 2.51237e-26 (rank : 25) | NC score | 0.789127 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
PDE1C_MOUSE
|
||||||
| θ value | 2.51237e-26 (rank : 26) | NC score | 0.788287 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
PDE1B_MOUSE
|
||||||
| θ value | 4.28545e-26 (rank : 27) | NC score | 0.799612 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01065, O35384 | Gene names | Pde1b, Pde1b1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE1B_HUMAN
|
||||||
| θ value | 9.54697e-26 (rank : 28) | NC score | 0.796338 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01064, Q92825 | Gene names | PDE1B, PDE1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE8A_HUMAN
|
||||||
| θ value | 1.05554e-24 (rank : 29) | NC score | 0.784989 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O60658, Q969I1, Q96PC9, Q96PD0, Q96PD1, Q9UMB7 | Gene names | PDE8A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17). | |||||
|
PDE7B_MOUSE
|
||||||
| θ value | 4.01107e-24 (rank : 30) | NC score | 0.812390 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QXQ1 | Gene names | Pde7b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE8A_MOUSE
|
||||||
| θ value | 8.93572e-24 (rank : 31) | NC score | 0.779952 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88502 | Gene names | Pde8a, Pde8 | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17) (MMPDE8). | |||||
|
PDE7B_HUMAN
|
||||||
| θ value | 1.99067e-23 (rank : 32) | NC score | 0.809437 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NP56 | Gene names | PDE7B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE8B_HUMAN
|
||||||
| θ value | 5.79196e-23 (rank : 33) | NC score | 0.777284 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95263, Q86XK8, Q8IUJ7, Q8IUJ8, Q8IUJ9, Q8IUK0, Q8N3T2 | Gene names | PDE8B | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8B (EC 3.1.4.17) (HSPDE8B). | |||||
|
PDE7A_HUMAN
|
||||||
| θ value | 6.40375e-22 (rank : 34) | NC score | 0.811582 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13946, O15380 | Gene names | PDE7A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (HCP1) (TM22). | |||||
|
PDE7A_MOUSE
|
||||||
| θ value | 5.42112e-21 (rank : 35) | NC score | 0.807829 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70453, Q9ERB3 | Gene names | Pde7a | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (P2A). | |||||
|
PDE3A_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 36) | NC score | 0.730039 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14432, O60865, Q13348 | Gene names | PDE3A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PDE3A_MOUSE
|
||||||
| θ value | 2.60593e-15 (rank : 37) | NC score | 0.732570 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Z0X4 | Gene names | Pde3a | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PDE3B_HUMAN
|
||||||
| θ value | 1.42992e-13 (rank : 38) | NC score | 0.733459 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13370, O00639, Q14408 | Gene names | PDE3B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase B (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase B) (CGI-PDE B) (CGIPDE1) (CGIP1). | |||||
|
VPP2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 39) | NC score | 0.024216 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15920, Q3U2X3, Q9JHJ2 | Gene names | Atp6v0a2, Atp6n1b, Tj6 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar proton translocating ATPase 116 kDa subunit a isoform 2 (V- ATPase 116 kDa isoform a2) (Immune suppressor factor J6B7). | |||||
|
ANGP2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.005291 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35608, Q3U1A1, Q9D2D2 | Gene names | Angpt2, Agpt2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-2 precursor (ANG-2). | |||||
|
VPP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.015579 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y487 | Gene names | ATP6V0A2 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar proton translocating ATPase 116 kDa subunit a isoform 2 (V- ATPase 116 kDa isoform a2) (TJ6). | |||||
|
NUDC3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.012955 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IVD9, Q9BTI3, Q9H7W9, Q9UPT4 | Gene names | NUDCD3, KIAA1068 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NudC domain-containing protein 3. | |||||
|
MOD5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.012018 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H3H1, Q6IAC9, Q96FJ3, Q96L45, Q9NXT7 | Gene names | TRIT1, IPT, MOD5 | |||
|
Domain Architecture |
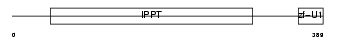
|
|||||
| Description | tRNA isopentenyltransferase, mitochondrial precursor (EC 2.5.1.8) (Isopentenyl-diphosphate:tRNA isopentenyltransferase) (IPP transferase) (IPTase) (IPPT) (hGRO1). | |||||
|
HIP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.000090 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
TLN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.008147 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.008106 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TXND3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.010229 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q715T0, Q80W74 | Gene names | Txndc3, Sptrx2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 3 (Spermatid-specific thioredoxin-2) (Sptrx-2). | |||||
|
HMMR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.002833 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
USH1C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.004276 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6N9, Q96B29, Q9UM04, Q9UM17, Q9UPC3 | Gene names | USH1C, AIE75 | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein) (Autoimmune enteropathy- related antigen AIE-75) (Antigen NY-CO-38/NY-CO-37) (PDZ-73 protein) (NY-REN-3 antigen). | |||||
|
USH1C_MOUSE
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.003641 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
ATR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.006777 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13535, Q7KYL3, Q93051, Q9BXK4 | Gene names | ATR, FRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein) (FRAP-related protein 1). | |||||
|
ATR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.006790 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JKK8 | Gene names | Atr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein). | |||||
|
CP24A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.000594 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64441 | Gene names | Cyp24a1, Cyp-24, Cyp24 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 24A1, mitochondrial precursor (EC 1.14.-.-) (P450- CC24) (Vitamin D(3) 24-hydroxylase) (1,25-dihydroxyvitamin D(3) 24- hydroxylase) (24-OHase). | |||||
|
GRIK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.000930 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13002 | Gene names | GRIK2, GLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 2 precursor (Glutamate receptor 6) (GluR-6) (GluR6) (Excitatory amino acid receptor 4) (EAA4). | |||||
|
HPLN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.002010 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QUP5, Q9D1G9, Q9Z1X7 | Gene names | Hapln1, Crtl1 | |||
|
Domain Architecture |
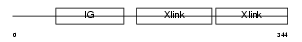
|
|||||
| Description | Hyaluronan and proteoglycan link protein 1 precursor (Proteoglycan link protein) (Cartilage link protein) (LP). | |||||
|
NEST_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.003766 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
PDE6C_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P51160, Q5VY29 | Gene names | PDE6C, PDEA2 | |||
|
Domain Architecture |

|
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE6C_MOUSE
|
||||||
| NC score | 0.999359 (rank : 2) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q91ZQ1, Q8R0D4 | Gene names | Pde6c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PDE6B_HUMAN
|
||||||
| NC score | 0.997365 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35913, Q9BWH5 | Gene names | PDE6B, PDEB | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase beta-subunit precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PDE6B_MOUSE
|
||||||
| NC score | 0.997253 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P23440 | Gene names | Pde6b, Mpb, Pdeb, rd | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PDE6A_MOUSE
|
||||||
| NC score | 0.996882 (rank : 5) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P27664 | Gene names | Pde6a, Mpa, Pdea | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha). | |||||
|
PDE6A_HUMAN
|
||||||
| NC score | 0.996767 (rank : 6) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16499 | Gene names | PDE6A, PDEA | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase alpha-subunit (EC 3.1.4.35) (GMP-PDE alpha) (PDE V-B1). | |||||
|
PDE11_HUMAN
|
||||||
| NC score | 0.963448 (rank : 7) | θ value | 1.29861e-115 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HCR9, Q53T16, Q96S76, Q9GZY7, Q9HB46, Q9NY45 | Gene names | PDE11A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE11_MOUSE
|
||||||
| NC score | 0.963381 (rank : 8) | θ value | 9.94316e-116 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P0C1Q2 | Gene names | Pde11a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual 3',5'-cyclic-AMP and -GMP phosphodiesterase 11A (EC 3.1.4.17) (EC 3.1.4.35) (cAMP and cGMP phosphodiesterase 11A). | |||||
|
PDE5A_MOUSE
|
||||||
| NC score | 0.960155 (rank : 9) | θ value | 8.73085e-104 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CG03 | Gene names | Pde5a, Pde5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE5A_HUMAN
|
||||||
| NC score | 0.959139 (rank : 10) | θ value | 1.26073e-102 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O76074, O75026, O75887, Q86UI0, Q86V66, Q9Y6Z6 | Gene names | PDE5A, PDE5 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-specific 3',5'-cyclic phosphodiesterase (EC 3.1.4.35) (CGB-PDE) (cGMP-binding cGMP-specific phosphodiesterase). | |||||
|
PDE10_HUMAN
|
||||||
| NC score | 0.944663 (rank : 11) | θ value | 1.27544e-62 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y233, Q9Y5T1 | Gene names | PDE10A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A (EC 3.1.4.17). | |||||
|
PDE2A_MOUSE
|
||||||
| NC score | 0.933088 (rank : 12) | θ value | 2.65329e-68 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q922S4, Q8K2U1 | Gene names | Pde2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE2A_HUMAN
|
||||||
| NC score | 0.931672 (rank : 13) | θ value | 3.4653e-68 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O00408 | Gene names | PDE2A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent 3',5'-cyclic phosphodiesterase (EC 3.1.4.17) (Cyclic GMP-stimulated phosphodiesterase) (CGS-PDE) (cGSPDE). | |||||
|
PDE9A_HUMAN
|
||||||
| NC score | 0.853302 (rank : 14) | θ value | 1.08979e-29 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O76083, O75490, O75491, O95225, Q86SF7, Q86SI6, Q86SJ3, Q86WN3, Q86WN4, Q86WN5, Q86WN6, Q86WN7, Q86WN8, Q86WN9, Q86WP0 | Gene names | PDE9A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE9A_MOUSE
|
||||||
| NC score | 0.851321 (rank : 15) | θ value | 1.02001e-27 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O70628 | Gene names | Pde9a, Pde8b | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A (EC 3.1.4.35). | |||||
|
PDE7B_MOUSE
|
||||||
| NC score | 0.812390 (rank : 16) | θ value | 4.01107e-24 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QXQ1 | Gene names | Pde7b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE7A_HUMAN
|
||||||
| NC score | 0.811582 (rank : 17) | θ value | 6.40375e-22 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13946, O15380 | Gene names | PDE7A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (HCP1) (TM22). | |||||
|
PDE4C_MOUSE
|
||||||
| NC score | 0.809858 (rank : 18) | θ value | 3.50572e-28 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3UEI1, Q8K0P4 | Gene names | Pde4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17). | |||||
|
PDE7B_HUMAN
|
||||||
| NC score | 0.809437 (rank : 19) | θ value | 1.99067e-23 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NP56 | Gene names | PDE7B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 7B (EC 3.1.4.17). | |||||
|
PDE4C_HUMAN
|
||||||
| NC score | 0.809267 (rank : 20) | θ value | 7.06379e-29 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08493, Q9UN44, Q9UN45, Q9UN46, Q9UPJ6 | Gene names | PDE4C | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17) (DPDE1) (PDE21). | |||||
|
PDE7A_MOUSE
|
||||||
| NC score | 0.807829 (rank : 21) | θ value | 5.42112e-21 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70453, Q9ERB3 | Gene names | Pde7a | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A (EC 3.1.4.17) (P2A). | |||||
|
PDE4D_MOUSE
|
||||||
| NC score | 0.807305 (rank : 22) | θ value | 2.77131e-33 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01063, Q6TRH9, Q8C4Q7, Q8CG05 | Gene names | Pde4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3). | |||||
|
PDE4A_MOUSE
|
||||||
| NC score | 0.806643 (rank : 23) | θ value | 2.77131e-33 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O89084, Q8R078, Q9JHQ4, Q9QX48, Q9QX49, Q9QXI8 | Gene names | Pde4a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17). | |||||
|
PDE4D_HUMAN
|
||||||
| NC score | 0.806558 (rank : 24) | θ value | 1.6247e-33 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08499, O43433, Q13549, Q13550, Q13551, Q7Z2L8, Q8IV84, Q8IVA9, Q8IVD2, Q8IVD3, Q9HCX7 | Gene names | PDE4D | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3) (PDE43). | |||||
|
PDE4A_HUMAN
|
||||||
| NC score | 0.806304 (rank : 25) | θ value | 2.12192e-33 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27815, O75522, O76092, Q16255, Q16691, Q8WUQ3 | Gene names | PDE4A | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17) (DPDE2) (PDE46). | |||||
|
PDE1B_MOUSE
|
||||||
| NC score | 0.799612 (rank : 26) | θ value | 4.28545e-26 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01065, O35384 | Gene names | Pde1b, Pde1b1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE1A_MOUSE
|
||||||
| NC score | 0.798865 (rank : 27) | θ value | 8.63488e-27 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61481, O35388 | Gene names | Pde1a | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE). | |||||
|
PDE4B_HUMAN
|
||||||
| NC score | 0.798275 (rank : 28) | θ value | 1.57365e-28 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q07343, O15443 | Gene names | PDE4B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4B (EC 3.1.4.17) (DPDE4) (PDE32). | |||||
|
PDE1A_HUMAN
|
||||||
| NC score | 0.796528 (rank : 29) | θ value | 2.51237e-26 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P54750, Q86VZ0, Q9C0K8, Q9C0K9, Q9C0L0, Q9C0L1, Q9C0L2, Q9C0L3, Q9C0L4, Q9UFX3 | Gene names | PDE1A | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1A (EC 3.1.4.17) (Cam-PDE 1A) (61 kDa Cam-PDE) (hCam-1). | |||||
|
PDE1B_HUMAN
|
||||||
| NC score | 0.796338 (rank : 30) | θ value | 9.54697e-26 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01064, Q92825 | Gene names | PDE1B, PDE1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B (EC 3.1.4.17) (Cam-PDE 1B) (63 kDa Cam-PDE). | |||||
|
PDE1C_HUMAN
|
||||||
| NC score | 0.789127 (rank : 31) | θ value | 2.51237e-26 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
PDE1C_MOUSE
|
||||||
| NC score | 0.788287 (rank : 32) | θ value | 2.51237e-26 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
PDE8A_HUMAN
|
||||||
| NC score | 0.784989 (rank : 33) | θ value | 1.05554e-24 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O60658, Q969I1, Q96PC9, Q96PD0, Q96PD1, Q9UMB7 | Gene names | PDE8A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17). | |||||
|
PDE8A_MOUSE
|
||||||
| NC score | 0.779952 (rank : 34) | θ value | 8.93572e-24 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88502 | Gene names | Pde8a, Pde8 | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17) (MMPDE8). | |||||
|
PDE8B_HUMAN
|
||||||
| NC score | 0.777284 (rank : 35) | θ value | 5.79196e-23 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95263, Q86XK8, Q8IUJ7, Q8IUJ8, Q8IUJ9, Q8IUK0, Q8N3T2 | Gene names | PDE8B | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8B (EC 3.1.4.17) (HSPDE8B). | |||||
|
PDE3B_HUMAN
|
||||||
| NC score | 0.733459 (rank : 36) | θ value | 1.42992e-13 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13370, O00639, Q14408 | Gene names | PDE3B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase B (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase B) (CGI-PDE B) (CGIPDE1) (CGIP1). | |||||
|
PDE3A_MOUSE
|
||||||
| NC score | 0.732570 (rank : 37) | θ value | 2.60593e-15 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Z0X4 | Gene names | Pde3a | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
PDE3A_HUMAN
|
||||||
| NC score | 0.730039 (rank : 38) | θ value | 4.02038e-16 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14432, O60865, Q13348 | Gene names | PDE3A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase A (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase A) (CGI-PDE A). | |||||
|
VPP2_MOUSE
|
||||||
| NC score | 0.024216 (rank : 39) | θ value | 0.21417 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15920, Q3U2X3, Q9JHJ2 | Gene names | Atp6v0a2, Atp6n1b, Tj6 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar proton translocating ATPase 116 kDa subunit a isoform 2 (V- ATPase 116 kDa isoform a2) (Immune suppressor factor J6B7). | |||||
|
VPP2_HUMAN
|
||||||
| NC score | 0.015579 (rank : 40) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y487 | Gene names | ATP6V0A2 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar proton translocating ATPase 116 kDa subunit a isoform 2 (V- ATPase 116 kDa isoform a2) (TJ6). | |||||
|
NUDC3_HUMAN
|
||||||
| NC score | 0.012955 (rank : 41) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IVD9, Q9BTI3, Q9H7W9, Q9UPT4 | Gene names | NUDCD3, KIAA1068 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NudC domain-containing protein 3. | |||||
|
MOD5_HUMAN
|
||||||
| NC score | 0.012018 (rank : 42) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H3H1, Q6IAC9, Q96FJ3, Q96L45, Q9NXT7 | Gene names | TRIT1, IPT, MOD5 | |||
|
Domain Architecture |

|
|||||
| Description | tRNA isopentenyltransferase, mitochondrial precursor (EC 2.5.1.8) (Isopentenyl-diphosphate:tRNA isopentenyltransferase) (IPP transferase) (IPTase) (IPPT) (hGRO1). | |||||
|
TXND3_MOUSE
|
||||||
| NC score | 0.010229 (rank : 43) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q715T0, Q80W74 | Gene names | Txndc3, Sptrx2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 3 (Spermatid-specific thioredoxin-2) (Sptrx-2). | |||||
|
TLN1_HUMAN
|
||||||
| NC score | 0.008147 (rank : 44) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN1_MOUSE
|
||||||
| NC score | 0.008106 (rank : 45) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
ATR_MOUSE
|
||||||
| NC score | 0.006790 (rank : 46) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JKK8 | Gene names | Atr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein). | |||||
|
ATR_HUMAN
|
||||||
| NC score | 0.006777 (rank : 47) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13535, Q7KYL3, Q93051, Q9BXK4 | Gene names | ATR, FRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein) (FRAP-related protein 1). | |||||
|
ANGP2_MOUSE
|
||||||
| NC score | 0.005291 (rank : 48) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35608, Q3U1A1, Q9D2D2 | Gene names | Angpt2, Agpt2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-2 precursor (ANG-2). | |||||
|
USH1C_HUMAN
|
||||||
| NC score | 0.004276 (rank : 49) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6N9, Q96B29, Q9UM04, Q9UM17, Q9UPC3 | Gene names | USH1C, AIE75 | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein) (Autoimmune enteropathy- related antigen AIE-75) (Antigen NY-CO-38/NY-CO-37) (PDZ-73 protein) (NY-REN-3 antigen). | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.003766 (rank : 50) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
USH1C_MOUSE
|
||||||
| NC score | 0.003641 (rank : 51) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
HMMR_MOUSE
|
||||||
| NC score | 0.002833 (rank : 52) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
HPLN1_MOUSE
|
||||||
| NC score | 0.002010 (rank : 53) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QUP5, Q9D1G9, Q9Z1X7 | Gene names | Hapln1, Crtl1 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 1 precursor (Proteoglycan link protein) (Cartilage link protein) (LP). | |||||
|
GRIK2_HUMAN
|
||||||
| NC score | 0.000930 (rank : 54) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13002 | Gene names | GRIK2, GLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 2 precursor (Glutamate receptor 6) (GluR-6) (GluR6) (Excitatory amino acid receptor 4) (EAA4). | |||||
|
CP24A_MOUSE
|
||||||
| NC score | 0.000594 (rank : 55) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64441 | Gene names | Cyp24a1, Cyp-24, Cyp24 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 24A1, mitochondrial precursor (EC 1.14.-.-) (P450- CC24) (Vitamin D(3) 24-hydroxylase) (1,25-dihydroxyvitamin D(3) 24- hydroxylase) (24-OHase). | |||||
|
HIP1_HUMAN
|
||||||
| NC score | 0.000090 (rank : 56) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||