Please be patient as the page loads
|
ATR_HUMAN
|
||||||
| SwissProt Accessions | Q13535, Q7KYL3, Q93051, Q9BXK4 | Gene names | ATR, FRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein) (FRAP-related protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ATR_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q13535, Q7KYL3, Q93051, Q9BXK4 | Gene names | ATR, FRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein) (FRAP-related protein 1). | |||||
|
ATR_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.996814 (rank : 2) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JKK8 | Gene names | Atr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein). | |||||
|
ATM_MOUSE
|
||||||
| θ value | 2.23575e-83 (rank : 3) | NC score | 0.800833 (rank : 3) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62388 | Gene names | Atm | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated homolog) (A-T, mutated homolog). | |||||
|
ATM_HUMAN
|
||||||
| θ value | 1.7238e-59 (rank : 4) | NC score | 0.794003 (rank : 4) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13315, O15429, Q12758, Q16551, Q93007, Q9NP02, Q9UCX7 | Gene names | ATM | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated) (A-T, mutated). | |||||
|
FRAP_MOUSE
|
||||||
| θ value | 3.84914e-51 (rank : 5) | NC score | 0.765309 (rank : 5) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JLN9, Q811J5, Q9CST1 | Gene names | Frap1, Frap | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
FRAP_HUMAN
|
||||||
| θ value | 1.91031e-50 (rank : 6) | NC score | 0.763938 (rank : 6) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
SMG1_MOUSE
|
||||||
| θ value | 4.40402e-47 (rank : 7) | NC score | 0.714042 (rank : 7) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
SMG1_HUMAN
|
||||||
| θ value | 5.7518e-47 (rank : 8) | NC score | 0.708641 (rank : 8) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
PRKDC_HUMAN
|
||||||
| θ value | 9.83387e-39 (rank : 9) | NC score | 0.693285 (rank : 10) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
PRKDC_MOUSE
|
||||||
| θ value | 1.57e-36 (rank : 10) | NC score | 0.696522 (rank : 9) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97313, O88187, P97928, Q307W9, Q3V2W8, Q8C2A7, Q9Z341 | Gene names | Prkdc, Xrcc7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (P460). | |||||
|
TRRAP_MOUSE
|
||||||
| θ value | 8.65492e-19 (rank : 11) | NC score | 0.477974 (rank : 11) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80YV3, Q8C0Z5, Q8K104 | Gene names | Trrap | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (Tra1 homolog). | |||||
|
TRRAP_HUMAN
|
||||||
| θ value | 1.13037e-18 (rank : 12) | NC score | 0.468437 (rank : 12) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
PK3C3_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 13) | NC score | 0.464131 (rank : 13) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6PF93, Q8R3S8 | Gene names | Pik3c3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3). | |||||
|
PK3C3_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 14) | NC score | 0.461052 (rank : 14) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NEB9, Q15134 | Gene names | PIK3C3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3) (Phosphatidylinositol 3-kinase p100 subunit). | |||||
|
PK3CG_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 15) | NC score | 0.368325 (rank : 18) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JHG7 | Gene names | Pik3cg, Pi3kg1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
PK3CG_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 16) | NC score | 0.369032 (rank : 17) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P48736, Q8IV23, Q9BZC8 | Gene names | PIK3CG | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
PK3CB_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 17) | NC score | 0.367675 (rank : 20) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42338 | Gene names | PIK3CB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CB_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 18) | NC score | 0.367773 (rank : 19) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BTI9 | Gene names | Pik3cb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
P3C2A_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 19) | NC score | 0.286253 (rank : 29) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
P3C2A_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 20) | NC score | 0.288660 (rank : 28) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61194, Q61182 | Gene names | Pik3c2a, Cpk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha) (Cpk-m) (p170). | |||||
|
PI4KB_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 21) | NC score | 0.376881 (rank : 15) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UBF8, O15096, P78405, Q9BWR6 | Gene names | PIK4CB, PI4KB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta) (NPIK) (PI4K92). | |||||
|
PK3CD_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 22) | NC score | 0.358702 (rank : 22) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00329, O15445 | Gene names | PIK3CD | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CD_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 23) | NC score | 0.359761 (rank : 21) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O35904 | Gene names | Pik3cd | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PI4KB_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 24) | NC score | 0.373726 (rank : 16) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BKC8, Q8C146 | Gene names | Pik4cb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta). | |||||
|
P3C2G_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 25) | NC score | 0.324750 (rank : 24) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70167, O70168 | Gene names | Pik3c2g | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
PI4KA_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 26) | NC score | 0.333679 (rank : 23) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42356, Q7Z625, Q9UPG2 | Gene names | PIK4CA, PIK4 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase alpha (EC 2.7.1.67) (PI4-kinase alpha) (PtdIns-4-kinase alpha) (PI4K-alpha). | |||||
|
P3C2G_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 27) | NC score | 0.316835 (rank : 27) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75747 | Gene names | PIK3C2G | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
PK3CA_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 28) | NC score | 0.321949 (rank : 25) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42336, Q99762 | Gene names | PIK3CA | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
PK3CA_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 29) | NC score | 0.321687 (rank : 26) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42337 | Gene names | Pik3ca | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
P3C2B_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 30) | NC score | 0.255712 (rank : 30) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00750, O95666, Q5SW99 | Gene names | PIK3C2B | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing beta polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-beta) (PtdIns-3-kinase C2 beta) (PI3K-C2beta) (C2-PI3K). | |||||
|
NR5A2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 31) | NC score | 0.025852 (rank : 39) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P45448 | Gene names | Nr5a2, Lrh1 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR5A2 (Liver receptor homolog 1) (LRH-1). | |||||
|
CAND1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 32) | NC score | 0.064651 (rank : 31) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86VP6, O94918, Q6PIY4, Q8NDJ4, Q96JZ9, Q96T19, Q9BTC4, Q9H0G2, Q9P0H7, Q9UF85 | Gene names | CAND1, KIAA0829, TIP120, TIP120A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 1 (Cullin-associated and neddylation-dissociated protein 1) (p120 CAND1) (TBP-interacting protein TIP120A) (TBP-interacting protein of 120 kDa A). | |||||
|
CAND1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 33) | NC score | 0.060458 (rank : 32) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZQ38, Q6PFR0, Q9CV45 | Gene names | Cand1, D10Ertd516e, Kiaa0829 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 1 (Cullin-associated and neddylation-dissociated protein 1) (p120 CAND1). | |||||
|
SYYC_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.037522 (rank : 34) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54577, O43276, Q53EN1 | Gene names | YARS | |||
|
Domain Architecture |
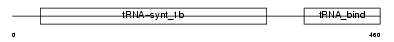
|
|||||
| Description | Tyrosyl-tRNA synthetase, cytoplasmic (EC 6.1.1.1) (Tyrosyl--tRNA ligase) (TyrRS). | |||||
|
CF152_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.029634 (rank : 37) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80ST9, Q9CYM9, Q9D5J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152 homolog. | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.019373 (rank : 41) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
SYYC_MOUSE
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.034088 (rank : 36) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WQ3, Q3TEC8, Q3U9A1, Q3UTA7, Q8BVT2, Q8C183 | Gene names | Yars | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosyl-tRNA synthetase, cytoplasmic (EC 6.1.1.1) (Tyrosyl--tRNA ligase) (TyrRS). | |||||
|
TBL3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.013778 (rank : 51) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12788 | Gene names | TBL3, SAZD | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein SAZD (Transducin beta-like 3 protein). | |||||
|
ABCC8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.011167 (rank : 56) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q09428, O75948, Q16583 | Gene names | ABCC8, SUR, SUR1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-binding cassette transporter sub-family C member 8 (Sulfonylurea receptor 1). | |||||
|
CF152_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.017758 (rank : 43) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
CTND1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.011437 (rank : 55) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60716, O15088, O60713, O60714, O60715, O60935, Q6RBX8, Q9UP71, Q9UP72, Q9UP73 | Gene names | CTNND1, KIAA0384 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
MK12_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | -0.000285 (rank : 69) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P53778, Q14260, Q99588, Q99672 | Gene names | MAPK12, ERK6, SAPK3 | |||
|
Domain Architecture |
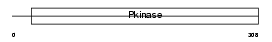
|
|||||
| Description | Mitogen-activated protein kinase 12 (EC 2.7.11.24) (Extracellular signal-regulated kinase 6) (ERK-6) (ERK5) (Stress-activated protein kinase 3) (Mitogen-activated protein kinase p38 gamma) (MAP kinase p38 gamma). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.009957 (rank : 57) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
TTBK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.009157 (rank : 59) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6IQ55, O94932, Q6ZN52, Q8IVV1 | Gene names | TTBK2, KIAA0847 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2 (EC 2.7.11.1). | |||||
|
ALS2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.015016 (rank : 49) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.008135 (rank : 62) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
INSC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.016766 (rank : 45) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q1MX18, Q1MX19, Q3C1V6, Q4AC95, Q4AC96, Q4AC97, Q4AC98 | Gene names | INSC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein inscuteable homolog. | |||||
|
KIFA3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.025892 (rank : 38) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92845, Q8NHU7, Q9H416 | Gene names | KIFAP3, SMAP | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-associated protein 3 (Smg GDS-associated protein). | |||||
|
PSMD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.019478 (rank : 40) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13200, Q12932, Q15321, Q96I12 | Gene names | PSMD2, TRAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 2 (26S proteasome regulatory subunit RPN1) (26S proteasome regulatory subunit S2) (26S proteasome subunit p97) (Tumor necrosis factor type 1 receptor- associated protein 2) (55.11 protein). | |||||
|
TCF25_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.016201 (rank : 46) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BQ70, Q2MK75, Q9UPV3 | Gene names | TCF25, KIAA1049, NULP1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 25 (Nuclear localized protein 1). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.019159 (rank : 42) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.015233 (rank : 48) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
UBD_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.035689 (rank : 35) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P63072, Q9WV10 | Gene names | Ubd, Fat10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin D (Ubiquitin-like protein FAT10) (Diubiquitin). | |||||
|
ULK4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.000722 (rank : 68) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3V129, Q2VP87, Q32LZ6, Q8BLS0, Q8C8Z5, Q9D4H6 | Gene names | Ulk4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK4 (EC 2.7.11.1) (Unc-51-like kinase 4). | |||||
|
ALS2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.013993 (rank : 50) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
CP250_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.005526 (rank : 65) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
ICAM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.009522 (rank : 58) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13597, Q61828 | Gene names | Icam1, Icam-1 | |||
|
Domain Architecture |

|
|||||
| Description | Intercellular adhesion molecule 1 precursor (ICAM-1) (MALA-2). | |||||
|
PARVG_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.015465 (rank : 47) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HBI0, Q9BQX5, Q9NSG1 | Gene names | PARVG | |||
|
Domain Architecture |
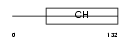
|
|||||
| Description | Gamma-parvin. | |||||
|
STK36_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | -0.001099 (rank : 70) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRP7, Q8TC32, Q9H9N9, Q9UF35, Q9ULE2 | Gene names | STK36, KIAA1278 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 36 (EC 2.7.11.1) (Fused homolog). | |||||
|
GATB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.012325 (rank : 54) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75879, Q9P0S6, Q9Y2B8 | Gene names | PET112L, PET112 | |||
|
Domain Architecture |

|
|||||
| Description | Probable glutamyl-tRNA(Gln) amidotransferase subunit B, mitochondrial precursor (EC 6.3.5.-) (Glu-ADT subunit B) (Cytochrome oxidase assembly factor PET112 homolog). | |||||
|
PDE6C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.006777 (rank : 63) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51160, Q5VY29 | Gene names | PDE6C, PDEA2 | |||
|
Domain Architecture |

|
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
PSMD2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.017455 (rank : 44) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VDM4 | Gene names | Psmd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 2 (26S proteasome regulatory subunit RPN1) (26S proteasome regulatory subunit S2) (26S proteasome subunit p97). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.012542 (rank : 53) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
ANR32_MOUSE
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.003182 (rank : 67) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R3P9, Q5XK30, Q8BHY5, Q921Y9 | Gene names | Ankrd32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 32. | |||||
|
CTND1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.008830 (rank : 60) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30999 | Gene names | Ctnnd1, Catns | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
GCN1L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.037719 (rank : 33) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92616, O95001, O95651, Q6P2S3, Q86X65, Q8N5I5, Q8WU80, Q99736, Q9UE60 | Gene names | GCN1L1, KIAA0219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GCN1-like protein 1 (HsGCN1). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.008136 (rank : 61) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
NEK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | -0.001659 (rank : 71) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
SMC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.013215 (rank : 52) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
SORT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.006091 (rank : 64) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PHU5, Q3UHE2, Q8K043, Q9QXW6 | Gene names | Sort1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sortilin precursor (Neurotensin receptor 3) (NTR3) (mNTR3). | |||||
|
TTBK2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.003375 (rank : 66) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
ATR_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q13535, Q7KYL3, Q93051, Q9BXK4 | Gene names | ATR, FRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein) (FRAP-related protein 1). | |||||
|
ATR_MOUSE
|
||||||
| NC score | 0.996814 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JKK8 | Gene names | Atr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein). | |||||
|
ATM_MOUSE
|
||||||
| NC score | 0.800833 (rank : 3) | θ value | 2.23575e-83 (rank : 3) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62388 | Gene names | Atm | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated homolog) (A-T, mutated homolog). | |||||
|
ATM_HUMAN
|
||||||
| NC score | 0.794003 (rank : 4) | θ value | 1.7238e-59 (rank : 4) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13315, O15429, Q12758, Q16551, Q93007, Q9NP02, Q9UCX7 | Gene names | ATM | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated) (A-T, mutated). | |||||
|
FRAP_MOUSE
|
||||||
| NC score | 0.765309 (rank : 5) | θ value | 3.84914e-51 (rank : 5) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JLN9, Q811J5, Q9CST1 | Gene names | Frap1, Frap | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
FRAP_HUMAN
|
||||||
| NC score | 0.763938 (rank : 6) | θ value | 1.91031e-50 (rank : 6) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
SMG1_MOUSE
|
||||||
| NC score | 0.714042 (rank : 7) | θ value | 4.40402e-47 (rank : 7) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
SMG1_HUMAN
|
||||||
| NC score | 0.708641 (rank : 8) | θ value | 5.7518e-47 (rank : 8) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
PRKDC_MOUSE
|
||||||
| NC score | 0.696522 (rank : 9) | θ value | 1.57e-36 (rank : 10) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97313, O88187, P97928, Q307W9, Q3V2W8, Q8C2A7, Q9Z341 | Gene names | Prkdc, Xrcc7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (P460). | |||||
|
PRKDC_HUMAN
|
||||||
| NC score | 0.693285 (rank : 10) | θ value | 9.83387e-39 (rank : 9) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
TRRAP_MOUSE
|
||||||
| NC score | 0.477974 (rank : 11) | θ value | 8.65492e-19 (rank : 11) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80YV3, Q8C0Z5, Q8K104 | Gene names | Trrap | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (Tra1 homolog). | |||||
|
TRRAP_HUMAN
|
||||||
| NC score | 0.468437 (rank : 12) | θ value | 1.13037e-18 (rank : 12) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
PK3C3_MOUSE
|
||||||
| NC score | 0.464131 (rank : 13) | θ value | 3.29651e-10 (rank : 13) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6PF93, Q8R3S8 | Gene names | Pik3c3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3). | |||||
|
PK3C3_HUMAN
|
||||||
| NC score | 0.461052 (rank : 14) | θ value | 1.25267e-09 (rank : 14) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NEB9, Q15134 | Gene names | PIK3C3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3) (Phosphatidylinositol 3-kinase p100 subunit). | |||||
|
PI4KB_HUMAN
|
||||||
| NC score | 0.376881 (rank : 15) | θ value | 2.44474e-05 (rank : 21) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UBF8, O15096, P78405, Q9BWR6 | Gene names | PIK4CB, PI4KB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta) (NPIK) (PI4K92). | |||||
|
PI4KB_MOUSE
|
||||||
| NC score | 0.373726 (rank : 16) | θ value | 3.19293e-05 (rank : 24) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BKC8, Q8C146 | Gene names | Pik4cb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta). | |||||
|
PK3CG_HUMAN
|
||||||
| NC score | 0.369032 (rank : 17) | θ value | 3.41135e-07 (rank : 16) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P48736, Q8IV23, Q9BZC8 | Gene names | PIK3CG | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
PK3CG_MOUSE
|
||||||
| NC score | 0.368325 (rank : 18) | θ value | 1.53129e-07 (rank : 15) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JHG7 | Gene names | Pik3cg, Pi3kg1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
PK3CB_MOUSE
|
||||||
| NC score | 0.367773 (rank : 19) | θ value | 1.09739e-05 (rank : 18) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BTI9 | Gene names | Pik3cb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CB_HUMAN
|
||||||
| NC score | 0.367675 (rank : 20) | θ value | 1.09739e-05 (rank : 17) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42338 | Gene names | PIK3CB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CD_MOUSE
|
||||||
| NC score | 0.359761 (rank : 21) | θ value | 2.44474e-05 (rank : 23) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O35904 | Gene names | Pik3cd | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CD_HUMAN
|
||||||
| NC score | 0.358702 (rank : 22) | θ value | 2.44474e-05 (rank : 22) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00329, O15445 | Gene names | PIK3CD | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PI4KA_HUMAN
|
||||||
| NC score | 0.333679 (rank : 23) | θ value | 0.000461057 (rank : 26) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42356, Q7Z625, Q9UPG2 | Gene names | PIK4CA, PIK4 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase alpha (EC 2.7.1.67) (PI4-kinase alpha) (PtdIns-4-kinase alpha) (PI4K-alpha). | |||||
|
P3C2G_MOUSE
|
||||||
| NC score | 0.324750 (rank : 24) | θ value | 0.000270298 (rank : 25) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70167, O70168 | Gene names | Pik3c2g | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
PK3CA_HUMAN
|
||||||
| NC score | 0.321949 (rank : 25) | θ value | 0.00175202 (rank : 28) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42336, Q99762 | Gene names | PIK3CA | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
PK3CA_MOUSE
|
||||||
| NC score | 0.321687 (rank : 26) | θ value | 0.00175202 (rank : 29) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42337 | Gene names | Pik3ca | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
P3C2G_HUMAN
|
||||||
| NC score | 0.316835 (rank : 27) | θ value | 0.000602161 (rank : 27) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75747 | Gene names | PIK3C2G | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
P3C2A_MOUSE
|
||||||
| NC score | 0.288660 (rank : 28) | θ value | 1.87187e-05 (rank : 20) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61194, Q61182 | Gene names | Pik3c2a, Cpk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha) (Cpk-m) (p170). | |||||
|
P3C2A_HUMAN
|
||||||
| NC score | 0.286253 (rank : 29) | θ value | 1.87187e-05 (rank : 19) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
P3C2B_HUMAN
|
||||||
| NC score | 0.255712 (rank : 30) | θ value | 0.0252991 (rank : 30) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00750, O95666, Q5SW99 | Gene names | PIK3C2B | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing beta polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-beta) (PtdIns-3-kinase C2 beta) (PI3K-C2beta) (C2-PI3K). | |||||
|
CAND1_HUMAN
|
||||||
| NC score | 0.064651 (rank : 31) | θ value | 0.0961366 (rank : 32) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86VP6, O94918, Q6PIY4, Q8NDJ4, Q96JZ9, Q96T19, Q9BTC4, Q9H0G2, Q9P0H7, Q9UF85 | Gene names | CAND1, KIAA0829, TIP120, TIP120A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 1 (Cullin-associated and neddylation-dissociated protein 1) (p120 CAND1) (TBP-interacting protein TIP120A) (TBP-interacting protein of 120 kDa A). | |||||
|
CAND1_MOUSE
|
||||||
| NC score | 0.060458 (rank : 32) | θ value | 0.21417 (rank : 33) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZQ38, Q6PFR0, Q9CV45 | Gene names | Cand1, D10Ertd516e, Kiaa0829 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 1 (Cullin-associated and neddylation-dissociated protein 1) (p120 CAND1). | |||||
|
GCN1L_HUMAN
|
||||||
| NC score | 0.037719 (rank : 33) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92616, O95001, O95651, Q6P2S3, Q86X65, Q8N5I5, Q8WU80, Q99736, Q9UE60 | Gene names | GCN1L1, KIAA0219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GCN1-like protein 1 (HsGCN1). | |||||
|
SYYC_HUMAN
|
||||||
| NC score | 0.037522 (rank : 34) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54577, O43276, Q53EN1 | Gene names | YARS | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosyl-tRNA synthetase, cytoplasmic (EC 6.1.1.1) (Tyrosyl--tRNA ligase) (TyrRS). | |||||
|
UBD_MOUSE
|
||||||
| NC score | 0.035689 (rank : 35) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P63072, Q9WV10 | Gene names | Ubd, Fat10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin D (Ubiquitin-like protein FAT10) (Diubiquitin). | |||||
|
SYYC_MOUSE
|
||||||
| NC score | 0.034088 (rank : 36) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WQ3, Q3TEC8, Q3U9A1, Q3UTA7, Q8BVT2, Q8C183 | Gene names | Yars | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosyl-tRNA synthetase, cytoplasmic (EC 6.1.1.1) (Tyrosyl--tRNA ligase) (TyrRS). | |||||
|
CF152_MOUSE
|
||||||
| NC score | 0.029634 (rank : 37) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80ST9, Q9CYM9, Q9D5J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152 homolog. | |||||
|
KIFA3_HUMAN
|
||||||
| NC score | 0.025892 (rank : 38) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92845, Q8NHU7, Q9H416 | Gene names | KIFAP3, SMAP | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-associated protein 3 (Smg GDS-associated protein). | |||||
|
NR5A2_MOUSE
|
||||||
| NC score | 0.025852 (rank : 39) | θ value | 0.0736092 (rank : 31) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P45448 | Gene names | Nr5a2, Lrh1 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR5A2 (Liver receptor homolog 1) (LRH-1). | |||||
|
PSMD2_HUMAN
|
||||||
| NC score | 0.019478 (rank : 40) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13200, Q12932, Q15321, Q96I12 | Gene names | PSMD2, TRAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 2 (26S proteasome regulatory subunit RPN1) (26S proteasome regulatory subunit S2) (26S proteasome subunit p97) (Tumor necrosis factor type 1 receptor- associated protein 2) (55.11 protein). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.019373 (rank : 41) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.019159 (rank : 42) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
CF152_HUMAN
|
||||||
| NC score | 0.017758 (rank : 43) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
PSMD2_MOUSE
|
||||||
| NC score | 0.017455 (rank : 44) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VDM4 | Gene names | Psmd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 2 (26S proteasome regulatory subunit RPN1) (26S proteasome regulatory subunit S2) (26S proteasome subunit p97). | |||||
|
INSC_HUMAN
|
||||||
| NC score | 0.016766 (rank : 45) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q1MX18, Q1MX19, Q3C1V6, Q4AC95, Q4AC96, Q4AC97, Q4AC98 | Gene names | INSC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein inscuteable homolog. | |||||
|
TCF25_HUMAN
|
||||||
| NC score | 0.016201 (rank : 46) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BQ70, Q2MK75, Q9UPV3 | Gene names | TCF25, KIAA1049, NULP1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 25 (Nuclear localized protein 1). | |||||
|
PARVG_HUMAN
|
||||||
| NC score | 0.015465 (rank : 47) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HBI0, Q9BQX5, Q9NSG1 | Gene names | PARVG | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-parvin. | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.015233 (rank : 48) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
ALS2_MOUSE
|
||||||
| NC score | 0.015016 (rank : 49) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
|
ALS2_HUMAN
|
||||||
| NC score | 0.013993 (rank : 50) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
TBL3_HUMAN
|
||||||
| NC score | 0.013778 (rank : 51) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12788 | Gene names | TBL3, SAZD | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein SAZD (Transducin beta-like 3 protein). | |||||
|
SMC2_MOUSE
|
||||||
| NC score | 0.013215 (rank : 52) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.012542 (rank : 53) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
GATB_HUMAN
|
||||||
| NC score | 0.012325 (rank : 54) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75879, Q9P0S6, Q9Y2B8 | Gene names | PET112L, PET112 | |||
|
Domain Architecture |

|
|||||
| Description | Probable glutamyl-tRNA(Gln) amidotransferase subunit B, mitochondrial precursor (EC 6.3.5.-) (Glu-ADT subunit B) (Cytochrome oxidase assembly factor PET112 homolog). | |||||
|
CTND1_HUMAN
|
||||||
| NC score | 0.011437 (rank : 55) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60716, O15088, O60713, O60714, O60715, O60935, Q6RBX8, Q9UP71, Q9UP72, Q9UP73 | Gene names | CTNND1, KIAA0384 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
ABCC8_HUMAN
|
||||||
| NC score | 0.011167 (rank : 56) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q09428, O75948, Q16583 | Gene names | ABCC8, SUR, SUR1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-binding cassette transporter sub-family C member 8 (Sulfonylurea receptor 1). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.009957 (rank : 57) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
ICAM1_MOUSE
|
||||||
| NC score | 0.009522 (rank : 58) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13597, Q61828 | Gene names | Icam1, Icam-1 | |||
|
Domain Architecture |

|
|||||
| Description | Intercellular adhesion molecule 1 precursor (ICAM-1) (MALA-2). | |||||
|
TTBK2_HUMAN
|
||||||
| NC score | 0.009157 (rank : 59) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6IQ55, O94932, Q6ZN52, Q8IVV1 | Gene names | TTBK2, KIAA0847 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2 (EC 2.7.11.1). | |||||
|
CTND1_MOUSE
|
||||||
| NC score | 0.008830 (rank : 60) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30999 | Gene names | Ctnnd1, Catns | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.008136 (rank : 61) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.008135 (rank : 62) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
PDE6C_HUMAN
|
||||||
| NC score | 0.006777 (rank : 63) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51160, Q5VY29 | Gene names | PDE6C, PDEA2 | |||
|
Domain Architecture |

|
|||||
| Description | Cone cGMP-specific 3',5'-cyclic phosphodiesterase alpha'-subunit (EC 3.1.4.35) (cGMP phosphodiesterase 6C). | |||||
|
SORT_MOUSE
|
||||||
| NC score | 0.006091 (rank : 64) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PHU5, Q3UHE2, Q8K043, Q9QXW6 | Gene names | Sort1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sortilin precursor (Neurotensin receptor 3) (NTR3) (mNTR3). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.005526 (rank : 65) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
TTBK2_MOUSE
|
||||||
| NC score | 0.003375 (rank : 66) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
ANR32_MOUSE
|
||||||
| NC score | 0.003182 (rank : 67) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R3P9, Q5XK30, Q8BHY5, Q921Y9 | Gene names | Ankrd32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 32. | |||||
|
ULK4_MOUSE
|
||||||
| NC score | 0.000722 (rank : 68) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3V129, Q2VP87, Q32LZ6, Q8BLS0, Q8C8Z5, Q9D4H6 | Gene names | Ulk4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK4 (EC 2.7.11.1) (Unc-51-like kinase 4). | |||||
|
MK12_HUMAN
|
||||||
| NC score | -0.000285 (rank : 69) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P53778, Q14260, Q99588, Q99672 | Gene names | MAPK12, ERK6, SAPK3 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 12 (EC 2.7.11.24) (Extracellular signal-regulated kinase 6) (ERK-6) (ERK5) (Stress-activated protein kinase 3) (Mitogen-activated protein kinase p38 gamma) (MAP kinase p38 gamma). | |||||
|
STK36_HUMAN
|
||||||
| NC score | -0.001099 (rank : 70) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRP7, Q8TC32, Q9H9N9, Q9UF35, Q9ULE2 | Gene names | STK36, KIAA1278 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 36 (EC 2.7.11.1) (Fused homolog). | |||||
|
NEK1_MOUSE
|
||||||
| NC score | -0.001659 (rank : 71) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||