Please be patient as the page loads
|
ALS2_MOUSE
|
||||||
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ALS2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.993643 (rank : 2) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
ALS2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
|
HERC3_HUMAN
|
||||||
| θ value | 2.13179e-17 (rank : 3) | NC score | 0.413553 (rank : 25) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
TSGA2_HUMAN
|
||||||
| θ value | 6.85773e-16 (rank : 4) | NC score | 0.539833 (rank : 6) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WYR4 | Gene names | TSGA2, TSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific gene A2 protein (Male meiotic metaphase chromosome- associated acidic protein) (Meichroacidin). | |||||
|
HERC2_HUMAN
|
||||||
| θ value | 1.52774e-15 (rank : 5) | NC score | 0.410975 (rank : 27) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_MOUSE
|
||||||
| θ value | 1.52774e-15 (rank : 6) | NC score | 0.409578 (rank : 28) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
MORN1_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 7) | NC score | 0.522197 (rank : 13) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5T089, Q8WW30, Q9H852 | Gene names | MORN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 1. | |||||
|
RCBT1_MOUSE
|
||||||
| θ value | 2.88119e-14 (rank : 8) | NC score | 0.470831 (rank : 20) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6NXM2, Q8BTZ6, Q8BZV0 | Gene names | Rcbtb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 1 (Regulator of chromosome condensation and BTB domain-containing protein 1). | |||||
|
MORN3_MOUSE
|
||||||
| θ value | 1.09485e-13 (rank : 9) | NC score | 0.522301 (rank : 12) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8C5T4, Q8VE21, Q9D5H6 | Gene names | Morn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 3. | |||||
|
RPGR_MOUSE
|
||||||
| θ value | 1.09485e-13 (rank : 10) | NC score | 0.530443 (rank : 8) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
TSGA2_MOUSE
|
||||||
| θ value | 1.09485e-13 (rank : 11) | NC score | 0.522895 (rank : 11) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VIG3, Q9DAL5 | Gene names | Tsga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific gene A2 protein (Male meiotic metaphase chromosome- associated acidic protein) (Meichroacidin). | |||||
|
HERC5_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 12) | NC score | 0.402495 (rank : 32) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UII4 | Gene names | HERC5, CEB1, CEBP1 | |||
|
Domain Architecture |

|
|||||
| Description | HECT domain and RCC1-like domain-containing protein 5 (Cyclin-E- binding protein 1). | |||||
|
RCBT1_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 13) | NC score | 0.468412 (rank : 21) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NDN9, Q969U9 | Gene names | RCBTB1, CLLD7, E4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 1 (Regulator of chromosome condensation and BTB domain-containing protein 1) (Chronic lymphocytic leukemia deletion region gene 7 protein) (CLL deletion region gene 7 protein). | |||||
|
MORN3_HUMAN
|
||||||
| θ value | 3.18553e-13 (rank : 14) | NC score | 0.518737 (rank : 15) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PF18, Q86YQ9 | Gene names | MORN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 3. | |||||
|
RPGR_HUMAN
|
||||||
| θ value | 3.18553e-13 (rank : 15) | NC score | 0.526002 (rank : 9) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92834, O00702, O00737, Q8N5T6, Q93039, Q9HD29, Q9UMR1 | Gene names | RPGR, RP3, XLRP3 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator. | |||||
|
RCBT2_MOUSE
|
||||||
| θ value | 9.26847e-13 (rank : 16) | NC score | 0.478407 (rank : 18) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99LJ7, Q3TUA3, Q8BMG2 | Gene names | Rcbtb2, Chc1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 2 (Regulator of chromosome condensation and BTB domain-containing protein 2) (Chromosome condensation 1-like). | |||||
|
RCC2_HUMAN
|
||||||
| θ value | 2.0648e-12 (rank : 17) | NC score | 0.566415 (rank : 3) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9P258, Q8IVL9, Q9BSN6, Q9NPV8 | Gene names | RCC2, KIAA1470, TD60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2 (Telophase disk protein of 60 kDa) (RCC1-like protein TD- 60). | |||||
|
RCC2_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 18) | NC score | 0.566375 (rank : 4) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BK67, Q6ZPQ0 | Gene names | Rcc2, Kiaa1470 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2. | |||||
|
RCBT2_HUMAN
|
||||||
| θ value | 6.00763e-12 (rank : 19) | NC score | 0.478199 (rank : 19) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O95199 | Gene names | RCBTB2, CHC1L, RLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 2 (Regulator of chromosome condensation and BTB domain-containing protein 2) (Chromosome condensation 1-like) (CHC1-L) (RCC1-like G exchanging factor). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 2.98157e-11 (rank : 20) | NC score | 0.382619 (rank : 33) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
JPH2_MOUSE
|
||||||
| θ value | 2.98157e-11 (rank : 21) | NC score | 0.407928 (rank : 30) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
JPH3_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 22) | NC score | 0.422324 (rank : 22) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WXH2, Q8N471, Q9HDC3, Q9HDC4 | Gene names | JPH3, JP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
JPH3_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 23) | NC score | 0.422280 (rank : 23) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ET77, Q9EQZ2 | Gene names | Jph3, Jp3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
JPH1_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 24) | NC score | 0.413582 (rank : 24) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ET80, Q9EQZ3 | Gene names | Jph1, Jp1 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
SRGEF_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 25) | NC score | 0.543096 (rank : 5) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80YD6, Q9QXB7 | Gene names | Sergef, Delgef, Gnefr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
NEK8_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 26) | NC score | 0.130074 (rank : 58) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86SG6, Q2M1S6, Q8NDH1 | Gene names | NEK8, JCK, NEK12A | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek8 (EC 2.7.11.1) (Never in mitosis A-related kinase 8) (NimA-related protein kinase 8) (NIMA-related kinase 12a). | |||||
|
JPH1_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 27) | NC score | 0.411096 (rank : 26) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HDC5 | Gene names | JPH1, JP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
NEK8_MOUSE
|
||||||
| θ value | 1.25267e-09 (rank : 28) | NC score | 0.129103 (rank : 60) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91ZR4, Q3U498, Q9D685 | Gene names | Nek8, Jck | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek8 (EC 2.7.11.1) (Never in mitosis A-related kinase 8) (NimA-related protein kinase 8). | |||||
|
JPH4_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 29) | NC score | 0.409448 (rank : 29) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96JJ6, Q8ND44, Q96DQ0 | Gene names | JPH4, JPHL1, KIAA1831 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
JPH4_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 30) | NC score | 0.406751 (rank : 31) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80WT0, Q8BMI1 | Gene names | Jph4, Jphl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
SRGEF_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 31) | NC score | 0.537892 (rank : 7) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UGK8, Q9UGK9 | Gene names | SERGEF, DELGEF, GNEFR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
RCC1_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 32) | NC score | 0.525876 (rank : 10) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8VE37, Q3UDB6 | Gene names | Rcc1, Chc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of chromosome condensation (Chromosome condensation protein 1). | |||||
|
MYCB2_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 33) | NC score | 0.334643 (rank : 36) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
RCC1_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 34) | NC score | 0.520910 (rank : 14) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P18754 | Gene names | RCC1, CHC1 | |||
|
Domain Architecture |
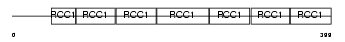
|
|||||
| Description | Regulator of chromosome condensation (Chromosome condensation protein 1) (Cell cycle regulatory protein). | |||||
|
WBS16_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 35) | NC score | 0.510561 (rank : 16) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96I51, Q548B1, Q9H0G7 | Gene names | WBSCR16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Williams-Beuren syndrome chromosome region 16 protein (RCC1-like G exchanging factor-like protein). | |||||
|
NEK9_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 36) | NC score | 0.129389 (rank : 59) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 900 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TD19, Q52LK6, Q8NCN0, Q8TCY4, Q9UPI4, Q9Y6S4, Q9Y6S5, Q9Y6S6 | Gene names | NEK9, KIAA1995, NEK8, NERCC | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek9 (EC 2.7.11.1) (NimA-related protein kinase 9) (Never in mitosis A-related kinase 9) (Nercc1 kinase) (NIMA-related kinase 8) (Nek8). | |||||
|
MYCB2_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 37) | NC score | 0.329283 (rank : 37) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
NEK9_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 38) | NC score | 0.130113 (rank : 57) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 890 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K1R7, Q8R3P1 | Gene names | Nek9, Nercc | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek9 (EC 2.7.11.1) (NimA-related protein kinase 9) (Never in mitosis A-related kinase 9). | |||||
|
WBS16_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 39) | NC score | 0.508186 (rank : 17) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CYF5 | Gene names | Wbscr16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Williams-Beuren syndrome chromosome region 16 protein homolog. | |||||
|
PKHF2_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 40) | NC score | 0.210493 (rank : 42) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H8W4 | Gene names | PLEKHF2, ZFYVE18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 2 (PH domain- containing family F member 2) (PH and FYVE domain-containing protein 2) (Phafin-2) (Zinc finger FYVE domain-containing protein 18). | |||||
|
PKHF2_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 41) | NC score | 0.208899 (rank : 44) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91WB4 | Gene names | Plekhf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 2 (PH domain- containing family F member 2). | |||||
|
ARHG6_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 42) | NC score | 0.111366 (rank : 69) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15052, Q15396, Q5JQ66, Q86XH0 | Gene names | ARHGEF6, COOL2, KIAA0006, PIXA | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 6 (Rac/Cdc42 guanine nucleotide exchange factor 6) (PAK-interacting exchange factor alpha) (Alpha-Pix) (COOL-2). | |||||
|
FGD2_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 43) | NC score | 0.157472 (rank : 49) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BY35, O88841, Q7TSE3, Q8VDH4 | Gene names | Fgd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 2. | |||||
|
FGD6_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 44) | NC score | 0.158432 (rank : 47) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
FARP1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 45) | NC score | 0.115022 (rank : 67) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
PKHF1_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 46) | NC score | 0.198089 (rank : 45) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3TB82, Q99M16 | Gene names | Plekhf1, Lapf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains). | |||||
|
ARHG6_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 47) | NC score | 0.101968 (rank : 73) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K4I3, Q8C9V4 | Gene names | Arhgef6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 6 (Rac/Cdc42 guanine nucleotide exchange factor 6). | |||||
|
ANKY1_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 48) | NC score | 0.210171 (rank : 43) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2S6, Q8IYX5, Q8NDK5, Q9NX10 | Gene names | ANKMY1, TSAL1, ZMYND13 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 1 (Testis-specific ankyrin-like protein 1) (Zinc-finger MYND domain-containing protein 13). | |||||
|
FGD6_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 49) | NC score | 0.157684 (rank : 48) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
PKHF1_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 50) | NC score | 0.191918 (rank : 46) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96S99, Q96K11, Q9BUB9 | Gene names | PLEKHF1, APPD, LAPF, ZFYVE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (PH and FYVE domain-containing protein 1) (Phafin-1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains) (Apoptosis-inducing protein) (Zinc finger FYVE domain-containing protein 15). | |||||
|
RIN3_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 51) | NC score | 0.139444 (rank : 52) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59729 | Gene names | Rin3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
RIN3_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 52) | NC score | 0.137665 (rank : 53) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TB24, Q8NF30, Q8TEE8, Q8WYP4, Q9H6A5, Q9HAG1 | Gene names | RIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
RIN2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 53) | NC score | 0.135463 (rank : 54) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WYP3, Q00425, Q5TFT8, Q9BQL3, Q9H071 | Gene names | RIN2, RASSF4 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 2 (Ras interaction/interference protein 2) (Ras inhibitor JC265) (Ras association domain family 4). | |||||
|
RIN2_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 54) | NC score | 0.134309 (rank : 56) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D684, Q99K06 | Gene names | Rin2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 2 (Ras interaction/interference protein 2). | |||||
|
ARHGA_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 55) | NC score | 0.112216 (rank : 68) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15013, O14665, Q68D55, Q8IWD9, Q8IY77 | Gene names | ARHGEF10, KIAA0294 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 10. | |||||
|
MORN2_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 56) | NC score | 0.368624 (rank : 34) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q502X0, Q6UL00 | Gene names | MORN2, MOPT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 2 (MORN motif protein in testis). | |||||
|
VAV2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 57) | NC score | 0.076104 (rank : 79) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P52735 | Gene names | VAV2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
FGD2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 58) | NC score | 0.140296 (rank : 50) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z6J4, Q5T8I1, Q6P6A8, Q6ZNL5, Q8IZ32, Q8N868, Q9H7M2 | Gene names | FGD2, ZFYVE4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 2 (Zinc finger FYVE domain-containing protein 4). | |||||
|
VAV2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 59) | NC score | 0.072159 (rank : 82) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60992 | Gene names | Vav2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
FARP2_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 60) | NC score | 0.107001 (rank : 71) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
FBX24_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 61) | NC score | 0.244277 (rank : 38) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75426, Q9H0G1 | Gene names | FBXO24, FBX24 | |||
|
Domain Architecture |
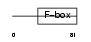
|
|||||
| Description | F-box only protein 24. | |||||
|
MORN2_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 62) | NC score | 0.350920 (rank : 35) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6UL01 | Gene names | Morn2, Mopt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 2 (MORN motif protein in testis). | |||||
|
FGD3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 63) | NC score | 0.126173 (rank : 61) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88842, Q8BQ72 | Gene names | Fgd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3. | |||||
|
RABX5_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 64) | NC score | 0.119802 (rank : 63) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UJ41 | Gene names | RABGEF1, RABEX5 | |||
|
Domain Architecture |

|
|||||
| Description | Rab5 GDP/GTP exchange factor (Rabex-5) (Rabaptin-5-associated exchange factor for Rab5). | |||||
|
RABX5_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 65) | NC score | 0.140202 (rank : 51) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JM13, Q3UIW0 | Gene names | Rabgef1, Rabex5 | |||
|
Domain Architecture |

|
|||||
| Description | Rab5 GDP/GTP exchange factor (Rabex-5). | |||||
|
SETD7_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 66) | NC score | 0.228579 (rank : 40) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WTS6, Q9C0E6 | Gene names | SETD7, KIAA1717, SET7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7) (Set9) (SET7/9). | |||||
|
SETD7_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 67) | NC score | 0.228766 (rank : 39) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VHL1 | Gene names | Setd7, Set7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7). | |||||
|
FBX24_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 68) | NC score | 0.220335 (rank : 41) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D417 | Gene names | Fbxo24, Fbx24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 24. | |||||
|
OGT1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 69) | NC score | 0.046099 (rank : 126) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
FGD3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 70) | NC score | 0.116375 (rank : 66) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5JSP0, Q7Z7D9, Q8N5G1 | Gene names | FGD3, ZFYVE5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3 (Zinc finger FYVE domain-containing protein 5). | |||||
|
FGD4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 71) | NC score | 0.134543 (rank : 55) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
ARHGA_MOUSE
|
||||||
| θ value | 0.21417 (rank : 72) | NC score | 0.098787 (rank : 74) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8C033, Q5DU38, Q80VH8, Q8BW76, Q922S7 | Gene names | Arhgef10, Kiaa0294 | |||
|
Domain Architecture |
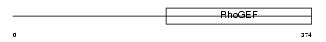
|
|||||
| Description | Rho guanine nucleotide exchange factor 10. | |||||
|
FGD1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 73) | NC score | 0.123219 (rank : 62) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
FGD1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 74) | NC score | 0.116870 (rank : 65) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
CP007_MOUSE
|
||||||
| θ value | 0.47712 (rank : 75) | NC score | 0.072473 (rank : 81) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C190, Q6PFD1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf7 homolog (5-day ovary-specific transcript 1 protein). | |||||
|
TR11B_MOUSE
|
||||||
| θ value | 0.47712 (rank : 76) | NC score | 0.030243 (rank : 127) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08712, O70202 | Gene names | Tnfrsf11b, Ocif, Opg | |||
|
Domain Architecture |
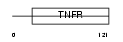
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11B precursor (Osteoprotegerin) (Osteoclastogenesis inhibitory factor). | |||||
|
KLH14_HUMAN
|
||||||
| θ value | 0.62314 (rank : 77) | NC score | 0.014514 (rank : 143) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P2G3 | Gene names | KLHL14, KIAA1384 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 14. | |||||
|
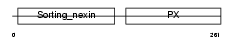
SNX1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 78) | NC score | 0.028557 (rank : 129) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13596, O60750, O60751 | Gene names | SNX1 | |||
|
Domain Architecture |
|
|||||
| Description | Sorting nexin-1. | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.008245 (rank : 154) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
GP112_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.008550 (rank : 153) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
CENA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.025002 (rank : 130) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75689 | Gene names | CENTA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 1 (Putative MAPK-activating protein PM25). | |||||
|
CSMD2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.005371 (rank : 159) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
HCFC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.017201 (rank : 137) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
HCFC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.017376 (rank : 136) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
PK3CD_MOUSE
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.011297 (rank : 149) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35904 | Gene names | Pik3cd | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
ATR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.015016 (rank : 141) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13535, Q7KYL3, Q93051, Q9BXK4 | Gene names | ATR, FRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein) (FRAP-related protein 1). | |||||
|
E2AK4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.012509 (rank : 145) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 984 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QZ05, Q6GQT4, Q6ZPT5, Q8C5S0, Q8CIF5, Q9CT30, Q9CUV9, Q9ESB6, Q9ESB7, Q9ESB8 | Gene names | Eif2ak4, Gcn2, Kiaa1338 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 2-alpha kinase 4 (EC 2.7.11.1) (GCN2-like protein) (mGCN2). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.011995 (rank : 146) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
PLEK_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.028667 (rank : 128) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
CEND1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.017891 (rank : 135) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
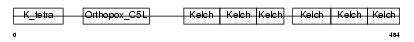
KLHL3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.008962 (rank : 152) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UH77, Q9UH75, Q9UH76, Q9ULU0, Q9Y6V6 | Gene names | KLHL3, KIAA1129 | |||
|
Domain Architecture |
|
|||||
| Description | Kelch-like protein 3. | |||||
|
MARK2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.010091 (rank : 150) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q05512, Q6PDR4, Q8BR95 | Gene names | Mark2, Emk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL Motif Kinase) (EMK1). | |||||
|
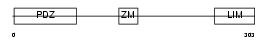
PDLI1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.005910 (rank : 158) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00151, Q5VZH5, Q9BPZ9 | Gene names | PDLIM1, CLIM1, CLP36 | |||
|
Domain Architecture |
|
|||||
| Description | PDZ and LIM domain protein 1 (Elfin) (LIM domain protein CLP-36) (C- terminal LIM domain protein 1). | |||||
|
PKHA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.024487 (rank : 131) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | -0.001744 (rank : 162) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CENA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.017934 (rank : 134) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NPF8, Q8N4Q6, Q96SD5 | Gene names | CENTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-alpha 2. | |||||
|
K0406_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.011444 (rank : 148) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91V83, Q9CSY0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0406 homolog. | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.005340 (rank : 160) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
POPD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.022614 (rank : 132) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HBU9, Q86UE7 | Gene names | POPDC2, POP2 | |||
|
Domain Architecture |

|
|||||
| Description | Popeye domain-containing protein 2 (Popeye protein 2). | |||||
|
PRAX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.007359 (rank : 157) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
CCNB3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.003688 (rank : 161) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CEND1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.015470 (rank : 140) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
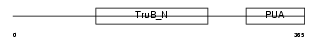
DKC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.009793 (rank : 151) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60832, O43845, Q96G67, Q9Y505 | Gene names | DKC1, NOLA4 | |||
|
Domain Architecture |
|
|||||
| Description | H/ACA ribonucleoprotein complex subunit 4 (EC 5.4.99.-) (Dyskerin) (Nucleolar protein family A member 4) (snoRNP protein DKC1) (Nopp140- associated protein of 57 kDa) (Nucleolar protein NAP57) (CBF5 homolog). | |||||
|
LPIN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.016133 (rank : 138) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92539 | Gene names | LPIN2, KIAA0249 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-2. | |||||
|
LPIN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.015679 (rank : 139) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99PI5, Q8C357, Q8C7I8, Q8CC85, Q8CHR7 | Gene names | Lpin2 | |||
|
Domain Architecture |
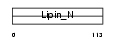
|
|||||
| Description | Lipin-2. | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.008070 (rank : 156) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
PANK4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.022441 (rank : 133) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NVE7, Q53EU3, Q5TA84, Q7RTX3, Q9H3X5 | Gene names | PANK4 | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 4 (EC 2.7.1.33) (Pantothenic acid kinase 4) (hPanK4). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.011837 (rank : 147) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
R113A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.008244 (rank : 155) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15541 | Gene names | RNF113A, RNF113, ZNF183 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113A (Zinc finger protein 183). | |||||
|
VKGC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.014476 (rank : 144) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38435, Q14415, Q6GU45 | Gene names | GGCX, GC | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent gamma-carboxylase (EC 6.4.-.-) (Gamma-glutamyl carboxylase) (Vitamin K gamma glutamyl carboxylase). | |||||
|
VKGC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.014577 (rank : 142) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QYC7, Q3UXN5, Q8CCB3 | Gene names | Ggcx | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent gamma-carboxylase (EC 6.4.-.-) (Gamma-glutamyl carboxylase) (Vitamin K gamma glutamyl carboxylase). | |||||
|
ARHG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.063997 (rank : 93) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NR80, Q9HDC6, Q9UPP0 | Gene names | ARHGEF4, KIAA1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
ARHG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.065499 (rank : 91) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TNR9, Q80TJ6 | Gene names | Arhgef4, Kiaa1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
ARHG5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.051662 (rank : 120) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12774 | Gene names | ARHGEF5, TIM | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 5 (Guanine nucleotide regulatory protein TIM) (Oncogene TIM) (p60 TIM) (Transforming immortalized mammary oncogene). | |||||
|
ARHG7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.065191 (rank : 92) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14155, Q6P9G3, Q6PII2, Q86W63, Q8N3M1 | Gene names | ARHGEF7, COOL1, KIAA0142, P85SPR, PAK3BP, PIXB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (COOL-1) (p85). | |||||
|
ARHG7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.060795 (rank : 96) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ES28, O08757, Q9ES27 | Gene names | Arhgef7, Pak3bp | |||
|
Domain Architecture |
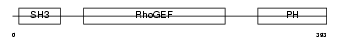
|
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (p85SPR). | |||||
|
ARHG9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.056996 (rank : 105) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43307, Q5JSL6 | Gene names | ARHGEF9, KIAA0424 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 9 (Rac/Cdc42 guanine nucleotide exchange factor 9) (Collybistin) (PEM-2 homolog). | |||||
|
ARHG9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.056533 (rank : 106) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3UTH8, Q3TQ60, Q80U06, Q8CAF9 | Gene names | Arhgef9, Kiaa0424 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 9 (Rac/Cdc42 guanine nucleotide exchange factor 9) (Collybistin). | |||||
|
ECT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.065910 (rank : 90) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H8V3, Q9NSV8, Q9NVW9 | Gene names | ECT2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
ECT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.066911 (rank : 89) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q07139 | Gene names | Ect2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
FARP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.087615 (rank : 75) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
FGD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.119702 (rank : 64) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96M96, Q6ULS2, Q8TCP6 | Gene names | FGD4, FRABP, ZFYVE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein) (Zinc finger FYVE domain-containing protein 6). | |||||
|
FGD5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.107637 (rank : 70) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
FGD5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.106697 (rank : 72) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80UZ0, Q8BHM5 | Gene names | Fgd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5. | |||||
|
GNRP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.050288 (rank : 123) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
GNRP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.052682 (rank : 118) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
HECD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.070181 (rank : 85) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5U5R9, Q5VZ97, Q5VZ99, Q8TCP5 | Gene names | HECTD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.069274 (rank : 86) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CDU6, Q8CBQ9 | Gene names | Hectd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.079897 (rank : 77) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T447, Q5T448, Q9H783 | Gene names | HECTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HECD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.079906 (rank : 76) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3U487, Q3TN76, Q641P3, Q8BQ74, Q8R1L6 | Gene names | Hectd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.055435 (rank : 108) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.056119 (rank : 107) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
ITCH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.053181 (rank : 114) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
ITCH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.053536 (rank : 113) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
KR202_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.057146 (rank : 104) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3LI61 | Gene names | KRTAP20-2, KAP20.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 20-2. | |||||
|
LORI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.073581 (rank : 80) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
LORI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.067412 (rank : 88) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
MCF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.050082 (rank : 125) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P10911, P14919, Q5JYJ4, Q8IUF3, Q8IUF4, Q9UJB3 | Gene names | MCF2, DBL | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene DBL (Proto-oncogene MCF-2) [Contains: MCF2-transforming protein; DBL-transforming protein]. | |||||
|
NGEF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.052048 (rank : 119) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N5V2, Q53QQ4, Q53ST7, Q6GMQ5, Q9NQD6 | Gene names | NGEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephexin-1 (Eph-interacting exchange protein) (Neuronal guanine nucleotide exchange factor). | |||||
|
NGEF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.050128 (rank : 124) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHT1, Q8R204, Q923H2, Q9JHT9 | Gene names | Ngef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephexin-1 (Eph-interacting exchange protein) (Neuronal guanine nucleotide exchange factor). | |||||
|
PKHG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.068120 (rank : 87) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ULL1, Q5T1F2 | Gene names | PLEKHG1, KIAA1209 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family G member 1. | |||||
|
PREX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.076565 (rank : 78) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TCU6, Q5JS95, Q5JS96, Q69YL2, Q7Z2L9, Q9BQH0, Q9BX55, Q9H4Q6, Q9P2D2, Q9UGQ4 | Gene names | PREX1, KIAA1415 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (P-Rex1 protein). | |||||
|
RIN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.058954 (rank : 101) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13671, O15010, Q00427, Q96CC8 | Gene names | RIN1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 1 (Ras interaction/interference protein 1) (Ras inhibitor JC99). | |||||
|
RIN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.059971 (rank : 97) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q921Q7 | Gene names | Rin1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 1 (Ras interaction/interference protein 1). | |||||
|
SMUF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.054315 (rank : 111) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
SMUF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.054044 (rank : 112) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
SMUF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.054377 (rank : 110) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.061450 (rank : 95) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
UBE3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.071376 (rank : 83) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
UBE3A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.071045 (rank : 84) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08759, P97482 | Gene names | Ube3a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (Oncogenic protein- associated protein E6-AP). | |||||
|
UBE3C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.059303 (rank : 98) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UBE3C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.059126 (rank : 99) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
WWP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.050586 (rank : 122) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
WWP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.050949 (rank : 121) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
WWP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.052888 (rank : 117) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
WWP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.053052 (rank : 116) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
ZFY16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.053169 (rank : 115) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z3T8, O15023, Q7LAU7, Q86T69, Q8N5L3, Q8NEK3 | Gene names | ZFYVE16, KIAA0305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 16 (Endofin) (Endosome- associated FYVE domain protein). | |||||
|
ZFY16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.054869 (rank : 109) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80U44, Q8BRD2, Q8CG97 | Gene names | Zfyve16, Kiaa0305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 16 (Endofin) (Endosomal- associated FYVE domain protein). | |||||
|
ZFY21_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.057552 (rank : 103) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BQ24, Q86T05, Q96LT1 | Gene names | ZFYVE21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 21. | |||||
|
ZFY21_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.058666 (rank : 102) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VCM3, Q9D1E2 | Gene names | Zfyve21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 21. | |||||
|
ZFY28_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.063412 (rank : 94) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HCC9 | Gene names | ZFYVE28, KIAA1643 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 28. | |||||
|
ZFYV9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.058957 (rank : 100) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95405, Q9UNE1, Q9Y5R7 | Gene names | ZFYVE9, MADHIP, SARA, SMADIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 9 (Mothers against decapentaplegic homolog-interacting protein) (Madh-interacting protein) (Smad anchor for receptor activation) (Receptor activation anchor) (hSARA) (Novel serine protease) (NSP). | |||||
|
ALS2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
|
ALS2_HUMAN
|
||||||
| NC score | 0.993643 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
RCC2_HUMAN
|
||||||
| NC score | 0.566415 (rank : 3) | θ value | 2.0648e-12 (rank : 17) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9P258, Q8IVL9, Q9BSN6, Q9NPV8 | Gene names | RCC2, KIAA1470, TD60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2 (Telophase disk protein of 60 kDa) (RCC1-like protein TD- 60). | |||||
|
RCC2_MOUSE
|
||||||
| NC score | 0.566375 (rank : 4) | θ value | 2.69671e-12 (rank : 18) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BK67, Q6ZPQ0 | Gene names | Rcc2, Kiaa1470 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2. | |||||
|
SRGEF_MOUSE
|
||||||
| NC score | 0.543096 (rank : 5) | θ value | 2.52405e-10 (rank : 25) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80YD6, Q9QXB7 | Gene names | Sergef, Delgef, Gnefr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
TSGA2_HUMAN
|
||||||
| NC score | 0.539833 (rank : 6) | θ value | 6.85773e-16 (rank : 4) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WYR4 | Gene names | TSGA2, TSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific gene A2 protein (Male meiotic metaphase chromosome- associated acidic protein) (Meichroacidin). | |||||
|
SRGEF_HUMAN
|
||||||
| NC score | 0.537892 (rank : 7) | θ value | 8.11959e-09 (rank : 31) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UGK8, Q9UGK9 | Gene names | SERGEF, DELGEF, GNEFR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
RPGR_MOUSE
|
||||||
| NC score | 0.530443 (rank : 8) | θ value | 1.09485e-13 (rank : 10) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
RPGR_HUMAN
|
||||||
| NC score | 0.526002 (rank : 9) | θ value | 3.18553e-13 (rank : 15) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92834, O00702, O00737, Q8N5T6, Q93039, Q9HD29, Q9UMR1 | Gene names | RPGR, RP3, XLRP3 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator. | |||||
|
RCC1_MOUSE
|
||||||
| NC score | 0.525876 (rank : 10) | θ value | 1.80886e-08 (rank : 32) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8VE37, Q3UDB6 | Gene names | Rcc1, Chc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of chromosome condensation (Chromosome condensation protein 1). | |||||
|
TSGA2_MOUSE
|
||||||
| NC score | 0.522895 (rank : 11) | θ value | 1.09485e-13 (rank : 11) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VIG3, Q9DAL5 | Gene names | Tsga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific gene A2 protein (Male meiotic metaphase chromosome- associated acidic protein) (Meichroacidin). | |||||
|
MORN3_MOUSE
|
||||||
| NC score | 0.522301 (rank : 12) | θ value | 1.09485e-13 (rank : 9) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8C5T4, Q8VE21, Q9D5H6 | Gene names | Morn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 3. | |||||
|
MORN1_HUMAN
|
||||||
| NC score | 0.522197 (rank : 13) | θ value | 3.40345e-15 (rank : 7) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5T089, Q8WW30, Q9H852 | Gene names | MORN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 1. | |||||
|
RCC1_HUMAN
|
||||||
| NC score | 0.520910 (rank : 14) | θ value | 1.17247e-07 (rank : 34) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P18754 | Gene names | RCC1, CHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of chromosome condensation (Chromosome condensation protein 1) (Cell cycle regulatory protein). | |||||
|
MORN3_HUMAN
|
||||||
| NC score | 0.518737 (rank : 15) | θ value | 3.18553e-13 (rank : 14) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PF18, Q86YQ9 | Gene names | MORN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 3. | |||||
|
WBS16_HUMAN
|
||||||
| NC score | 0.510561 (rank : 16) | θ value | 1.17247e-07 (rank : 35) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96I51, Q548B1, Q9H0G7 | Gene names | WBSCR16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Williams-Beuren syndrome chromosome region 16 protein (RCC1-like G exchanging factor-like protein). | |||||
|
WBS16_MOUSE
|
||||||
| NC score | 0.508186 (rank : 17) | θ value | 4.45536e-07 (rank : 39) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CYF5 | Gene names | Wbscr16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Williams-Beuren syndrome chromosome region 16 protein homolog. | |||||
|
RCBT2_MOUSE
|
||||||
| NC score | 0.478407 (rank : 18) | θ value | 9.26847e-13 (rank : 16) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99LJ7, Q3TUA3, Q8BMG2 | Gene names | Rcbtb2, Chc1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 2 (Regulator of chromosome condensation and BTB domain-containing protein 2) (Chromosome condensation 1-like). | |||||
|
RCBT2_HUMAN
|
||||||
| NC score | 0.478199 (rank : 19) | θ value | 6.00763e-12 (rank : 19) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O95199 | Gene names | RCBTB2, CHC1L, RLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 2 (Regulator of chromosome condensation and BTB domain-containing protein 2) (Chromosome condensation 1-like) (CHC1-L) (RCC1-like G exchanging factor). | |||||
|
RCBT1_MOUSE
|
||||||
| NC score | 0.470831 (rank : 20) | θ value | 2.88119e-14 (rank : 8) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6NXM2, Q8BTZ6, Q8BZV0 | Gene names | Rcbtb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 1 (Regulator of chromosome condensation and BTB domain-containing protein 1). | |||||
|
RCBT1_HUMAN
|
||||||
| NC score | 0.468412 (rank : 21) | θ value | 1.86753e-13 (rank : 13) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NDN9, Q969U9 | Gene names | RCBTB1, CLLD7, E4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 1 (Regulator of chromosome condensation and BTB domain-containing protein 1) (Chronic lymphocytic leukemia deletion region gene 7 protein) (CLL deletion region gene 7 protein). | |||||
|
JPH3_HUMAN
|
||||||
| NC score | 0.422324 (rank : 22) | θ value | 3.89403e-11 (rank : 22) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WXH2, Q8N471, Q9HDC3, Q9HDC4 | Gene names | JPH3, JP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
JPH3_MOUSE
|
||||||
| NC score | 0.422280 (rank : 23) | θ value | 3.89403e-11 (rank : 23) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ET77, Q9EQZ2 | Gene names | Jph3, Jp3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
JPH1_MOUSE
|
||||||
| NC score | 0.413582 (rank : 24) | θ value | 1.9326e-10 (rank : 24) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ET80, Q9EQZ3 | Gene names | Jph1, Jp1 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
HERC3_HUMAN
|
||||||
| NC score | 0.413553 (rank : 25) | θ value | 2.13179e-17 (rank : 3) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
JPH1_HUMAN
|
||||||
| NC score | 0.411096 (rank : 26) | θ value | 1.25267e-09 (rank : 27) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HDC5 | Gene names | JPH1, JP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
HERC2_HUMAN
|
||||||
| NC score | 0.410975 (rank : 27) | θ value | 1.52774e-15 (rank : 5) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_MOUSE
|
||||||
| NC score | 0.409578 (rank : 28) | θ value | 1.52774e-15 (rank : 6) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
JPH4_HUMAN
|
||||||
| NC score | 0.409448 (rank : 29) | θ value | 2.79066e-09 (rank : 29) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96JJ6, Q8ND44, Q96DQ0 | Gene names | JPH4, JPHL1, KIAA1831 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
JPH2_MOUSE
|
||||||
| NC score | 0.407928 (rank : 30) | θ value | 2.98157e-11 (rank : 21) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
JPH4_MOUSE
|
||||||
| NC score | 0.406751 (rank : 31) | θ value | 2.79066e-09 (rank : 30) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80WT0, Q8BMI1 | Gene names | Jph4, Jphl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
HERC5_HUMAN
|
||||||
| NC score | 0.402495 (rank : 32) | θ value | 1.86753e-13 (rank : 12) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UII4 | Gene names | HERC5, CEB1, CEBP1 | |||
|
Domain Architecture |

|
|||||
| Description | HECT domain and RCC1-like domain-containing protein 5 (Cyclin-E- binding protein 1). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.382619 (rank : 33) | θ value | 2.98157e-11 (rank : 20) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
MORN2_HUMAN
|
||||||
| NC score | 0.368624 (rank : 34) | θ value | 0.00665767 (rank : 56) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q502X0, Q6UL00 | Gene names | MORN2, MOPT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 2 (MORN motif protein in testis). | |||||
|
MORN2_MOUSE
|
||||||
| NC score | 0.350920 (rank : 35) | θ value | 0.0148317 (rank : 62) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6UL01 | Gene names | Morn2, Mopt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 2 (MORN motif protein in testis). | |||||
|
MYCB2_HUMAN
|
||||||
| NC score | 0.334643 (rank : 36) | θ value | 3.08544e-08 (rank : 33) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
MYCB2_MOUSE
|
||||||
| NC score | 0.329283 (rank : 37) | θ value | 3.41135e-07 (rank : 37) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
FBX24_HUMAN
|
||||||
| NC score | 0.244277 (rank : 38) | θ value | 0.0148317 (rank : 61) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75426, Q9H0G1 | Gene names | FBXO24, FBX24 | |||
|
Domain Architecture |

|
|||||
| Description | F-box only protein 24. | |||||
|
SETD7_MOUSE
|
||||||
| NC score | 0.228766 (rank : 39) | θ value | 0.0252991 (rank : 67) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VHL1 | Gene names | Setd7, Set7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7). | |||||
|
SETD7_HUMAN
|
||||||
| NC score | 0.228579 (rank : 40) | θ value | 0.0252991 (rank : 66) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WTS6, Q9C0E6 | Gene names | SETD7, KIAA1717, SET7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7) (Set9) (SET7/9). | |||||
|
FBX24_MOUSE
|
||||||
| NC score | 0.220335 (rank : 41) | θ value | 0.0563607 (rank : 68) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D417 | Gene names | Fbxo24, Fbx24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 24. | |||||
|
PKHF2_HUMAN
|
||||||
| NC score | 0.210493 (rank : 42) | θ value | 8.40245e-06 (rank : 40) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H8W4 | Gene names | PLEKHF2, ZFYVE18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 2 (PH domain- containing family F member 2) (PH and FYVE domain-containing protein 2) (Phafin-2) (Zinc finger FYVE domain-containing protein 18). | |||||
|
ANKY1_HUMAN
|
||||||
| NC score | 0.210171 (rank : 43) | θ value | 0.000270298 (rank : 48) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2S6, Q8IYX5, Q8NDK5, Q9NX10 | Gene names | ANKMY1, TSAL1, ZMYND13 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 1 (Testis-specific ankyrin-like protein 1) (Zinc-finger MYND domain-containing protein 13). | |||||
|
PKHF2_MOUSE
|
||||||
| NC score | 0.208899 (rank : 44) | θ value | 1.09739e-05 (rank : 41) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91WB4 | Gene names | Plekhf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 2 (PH domain- containing family F member 2). | |||||
|
PKHF1_MOUSE
|
||||||
| NC score | 0.198089 (rank : 45) | θ value | 0.000158464 (rank : 46) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3TB82, Q99M16 | Gene names | Plekhf1, Lapf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains). | |||||
|
PKHF1_HUMAN
|
||||||
| NC score | 0.191918 (rank : 46) | θ value | 0.000461057 (rank : 50) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96S99, Q96K11, Q9BUB9 | Gene names | PLEKHF1, APPD, LAPF, ZFYVE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (PH and FYVE domain-containing protein 1) (Phafin-1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains) (Apoptosis-inducing protein) (Zinc finger FYVE domain-containing protein 15). | |||||
|
FGD6_HUMAN
|
||||||
| NC score | 0.158432 (rank : 47) | θ value | 9.29e-05 (rank : 44) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
FGD6_MOUSE
|
||||||
| NC score | 0.157684 (rank : 48) | θ value | 0.000270298 (rank : 49) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
FGD2_MOUSE
|
||||||
| NC score | 0.157472 (rank : 49) | θ value | 7.1131e-05 (rank : 43) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BY35, O88841, Q7TSE3, Q8VDH4 | Gene names | Fgd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 2. | |||||
|
FGD2_HUMAN
|
||||||
| NC score | 0.140296 (rank : 50) | θ value | 0.0113563 (rank : 58) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z6J4, Q5T8I1, Q6P6A8, Q6ZNL5, Q8IZ32, Q8N868, Q9H7M2 | Gene names | FGD2, ZFYVE4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 2 (Zinc finger FYVE domain-containing protein 4). | |||||
|
RABX5_MOUSE
|
||||||
| NC score | 0.140202 (rank : 51) | θ value | 0.0252991 (rank : 65) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JM13, Q3UIW0 | Gene names | Rabgef1, Rabex5 | |||
|
Domain Architecture |

|
|||||
| Description | Rab5 GDP/GTP exchange factor (Rabex-5). | |||||
|
RIN3_MOUSE
|
||||||
| NC score | 0.139444 (rank : 52) | θ value | 0.00102713 (rank : 51) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59729 | Gene names | Rin3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
RIN3_HUMAN
|
||||||
| NC score | 0.137665 (rank : 53) | θ value | 0.00228821 (rank : 52) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TB24, Q8NF30, Q8TEE8, Q8WYP4, Q9H6A5, Q9HAG1 | Gene names | RIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
RIN2_HUMAN
|
||||||
| NC score | 0.135463 (rank : 54) | θ value | 0.00390308 (rank : 53) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WYP3, Q00425, Q5TFT8, Q9BQL3, Q9H071 | Gene names | RIN2, RASSF4 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 2 (Ras interaction/interference protein 2) (Ras inhibitor JC265) (Ras association domain family 4). | |||||
|
FGD4_MOUSE
|
||||||
| NC score | 0.134543 (rank : 55) | θ value | 0.163984 (rank : 71) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
RIN2_MOUSE
|
||||||
| NC score | 0.134309 (rank : 56) | θ value | 0.00390308 (rank : 54) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D684, Q99K06 | Gene names | Rin2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 2 (Ras interaction/interference protein 2). | |||||
|
NEK9_MOUSE
|
||||||
| NC score | 0.130113 (rank : 57) | θ value | 4.45536e-07 (rank : 38) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 890 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K1R7, Q8R3P1 | Gene names | Nek9, Nercc | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek9 (EC 2.7.11.1) (NimA-related protein kinase 9) (Never in mitosis A-related kinase 9). | |||||
|
NEK8_HUMAN
|
||||||
| NC score | 0.130074 (rank : 58) | θ value | 5.62301e-10 (rank : 26) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86SG6, Q2M1S6, Q8NDH1 | Gene names | NEK8, JCK, NEK12A | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek8 (EC 2.7.11.1) (Never in mitosis A-related kinase 8) (NimA-related protein kinase 8) (NIMA-related kinase 12a). | |||||
|
NEK9_HUMAN
|
||||||
| NC score | 0.129389 (rank : 59) | θ value | 1.99992e-07 (rank : 36) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 900 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TD19, Q52LK6, Q8NCN0, Q8TCY4, Q9UPI4, Q9Y6S4, Q9Y6S5, Q9Y6S6 | Gene names | NEK9, KIAA1995, NEK8, NERCC | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek9 (EC 2.7.11.1) (NimA-related protein kinase 9) (Never in mitosis A-related kinase 9) (Nercc1 kinase) (NIMA-related kinase 8) (Nek8). | |||||
|
NEK8_MOUSE
|
||||||
| NC score | 0.129103 (rank : 60) | θ value | 1.25267e-09 (rank : 28) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91ZR4, Q3U498, Q9D685 | Gene names | Nek8, Jck | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek8 (EC 2.7.11.1) (Never in mitosis A-related kinase 8) (NimA-related protein kinase 8). | |||||
|
FGD3_MOUSE
|
||||||
| NC score | 0.126173 (rank : 61) | θ value | 0.0252991 (rank : 63) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88842, Q8BQ72 | Gene names | Fgd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3. | |||||
|
FGD1_HUMAN
|
||||||
| NC score | 0.123219 (rank : 62) | θ value | 0.21417 (rank : 73) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
RABX5_HUMAN
|
||||||
| NC score | 0.119802 (rank : 63) | θ value | 0.0252991 (rank : 64) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UJ41 | Gene names | RABGEF1, RABEX5 | |||
|
Domain Architecture |

|
|||||
| Description | Rab5 GDP/GTP exchange factor (Rabex-5) (Rabaptin-5-associated exchange factor for Rab5). | |||||
|
FGD4_HUMAN
|
||||||
| NC score | 0.119702 (rank : 64) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96M96, Q6ULS2, Q8TCP6 | Gene names | FGD4, FRABP, ZFYVE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein) (Zinc finger FYVE domain-containing protein 6). | |||||
|
FGD1_MOUSE
|
||||||
| NC score | 0.116870 (rank : 65) | θ value | 0.365318 (rank : 74) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
FGD3_HUMAN
|
||||||
| NC score | 0.116375 (rank : 66) | θ value | 0.125558 (rank : 70) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5JSP0, Q7Z7D9, Q8N5G1 | Gene names | FGD3, ZFYVE5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3 (Zinc finger FYVE domain-containing protein 5). | |||||
|
FARP1_HUMAN
|
||||||
| NC score | 0.115022 (rank : 67) | θ value | 0.000121331 (rank : 45) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
ARHGA_HUMAN
|
||||||
| NC score | 0.112216 (rank : 68) | θ value | 0.00665767 (rank : 55) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15013, O14665, Q68D55, Q8IWD9, Q8IY77 | Gene names | ARHGEF10, KIAA0294 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 10. | |||||
|
ARHG6_HUMAN
|
||||||
| NC score | 0.111366 (rank : 69) | θ value | 2.44474e-05 (rank : 42) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15052, Q15396, Q5JQ66, Q86XH0 | Gene names | ARHGEF6, COOL2, KIAA0006, PIXA | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 6 (Rac/Cdc42 guanine nucleotide exchange factor 6) (PAK-interacting exchange factor alpha) (Alpha-Pix) (COOL-2). | |||||
|
FGD5_HUMAN
|
||||||
| NC score | 0.107637 (rank : 70) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
FARP2_MOUSE
|
||||||
| NC score | 0.107001 (rank : 71) | θ value | 0.0148317 (rank : 60) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
FGD5_MOUSE
|
||||||
| NC score | 0.106697 (rank : 72) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80UZ0, Q8BHM5 | Gene names | Fgd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5. | |||||
|
ARHG6_MOUSE
|
||||||
| NC score | 0.101968 (rank : 73) | θ value | 0.00020696 (rank : 47) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K4I3, Q8C9V4 | Gene names | Arhgef6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 6 (Rac/Cdc42 guanine nucleotide exchange factor 6). | |||||
|
ARHGA_MOUSE
|
||||||
| NC score | 0.098787 (rank : 74) | θ value | 0.21417 (rank : 72) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8C033, Q5DU38, Q80VH8, Q8BW76, Q922S7 | Gene names | Arhgef10, Kiaa0294 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 10. | |||||
|
FARP2_HUMAN
|
||||||
| NC score | 0.087615 (rank : 75) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
HECD3_MOUSE
|
||||||
| NC score | 0.079906 (rank : 76) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3U487, Q3TN76, Q641P3, Q8BQ74, Q8R1L6 | Gene names | Hectd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HECD3_HUMAN
|
||||||
| NC score | 0.079897 (rank : 77) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T447, Q5T448, Q9H783 | Gene names | HECTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
PREX1_HUMAN
|
||||||
| NC score | 0.076565 (rank : 78) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TCU6, Q5JS95, Q5JS96, Q69YL2, Q7Z2L9, Q9BQH0, Q9BX55, Q9H4Q6, Q9P2D2, Q9UGQ4 | Gene names | PREX1, KIAA1415 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (P-Rex1 protein). | |||||
|
VAV2_HUMAN
|
||||||
| NC score | 0.076104 (rank : 79) | θ value | 0.00869519 (rank : 57) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P52735 | Gene names | VAV2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
LORI_HUMAN
|
||||||
| NC score | 0.073581 (rank : 80) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
CP007_MOUSE
|
||||||
| NC score | 0.072473 (rank : 81) | θ value | 0.47712 (rank : 75) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C190, Q6PFD1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf7 homolog (5-day ovary-specific transcript 1 protein). | |||||
|
VAV2_MOUSE
|
||||||
| NC score | 0.072159 (rank : 82) | θ value | 0.0113563 (rank : 59) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60992 | Gene names | Vav2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
UBE3A_HUMAN
|
||||||
| NC score | 0.071376 (rank : 83) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
UBE3A_MOUSE
|
||||||
| NC score | 0.071045 (rank : 84) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08759, P97482 | Gene names | Ube3a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (Oncogenic protein- associated protein E6-AP). | |||||
|
HECD2_HUMAN
|
||||||
| NC score | 0.070181 (rank : 85) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5U5R9, Q5VZ97, Q5VZ99, Q8TCP5 | Gene names | HECTD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD2_MOUSE
|
||||||
| NC score | 0.069274 (rank : 86) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CDU6, Q8CBQ9 | Gene names | Hectd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
PKHG1_HUMAN
|
||||||
| NC score | 0.068120 (rank : 87) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ULL1, Q5T1F2 | Gene names | PLEKHG1, KIAA1209 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family G member 1. | |||||
|
LORI_MOUSE
|
||||||
| NC score | 0.067412 (rank : 88) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
ECT2_MOUSE
|
||||||
| NC score | 0.066911 (rank : 89) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q07139 | Gene names | Ect2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
ECT2_HUMAN
|
||||||
| NC score | 0.065910 (rank : 90) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H8V3, Q9NSV8, Q9NVW9 | Gene names | ECT2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
ARHG4_MOUSE
|
||||||
| NC score | 0.065499 (rank : 91) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TNR9, Q80TJ6 | Gene names | Arhgef4, Kiaa1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
ARHG7_HUMAN
|
||||||
| NC score | 0.065191 (rank : 92) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14155, Q6P9G3, Q6PII2, Q86W63, Q8N3M1 | Gene names | ARHGEF7, COOL1, KIAA0142, P85SPR, PAK3BP, PIXB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (COOL-1) (p85). | |||||
|
ARHG4_HUMAN
|
||||||
| NC score | 0.063997 (rank : 93) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NR80, Q9HDC6, Q9UPP0 | Gene names | ARHGEF4, KIAA1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
ZFY28_HUMAN
|
||||||
| NC score | 0.063412 (rank : 94) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HCC9 | Gene names | ZFYVE28, KIAA1643 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 28. | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.061450 (rank : 95) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
ARHG7_MOUSE
|
||||||
| NC score | 0.060795 (rank : 96) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ES28, O08757, Q9ES27 | Gene names | Arhgef7, Pak3bp | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (p85SPR). | |||||
|
RIN1_MOUSE
|
||||||
| NC score | 0.059971 (rank : 97) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q921Q7 | Gene names | Rin1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 1 (Ras interaction/interference protein 1). | |||||
|
UBE3C_HUMAN
|
||||||
| NC score | 0.059303 (rank : 98) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UBE3C_MOUSE
|
||||||
| NC score | 0.059126 (rank : 99) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
ZFYV9_HUMAN
|
||||||
| NC score | 0.058957 (rank : 100) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95405, Q9UNE1, Q9Y5R7 | Gene names | ZFYVE9, MADHIP, SARA, SMADIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 9 (Mothers against decapentaplegic homolog-interacting protein) (Madh-interacting protein) (Smad anchor for receptor activation) (Receptor activation anchor) (hSARA) (Novel serine protease) (NSP). | |||||
|
RIN1_HUMAN
|
||||||
| NC score | 0.058954 (rank : 101) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13671, O15010, Q00427, Q96CC8 | Gene names | RIN1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 1 (Ras interaction/interference protein 1) (Ras inhibitor JC99). | |||||
|
ZFY21_MOUSE
|
||||||
| NC score | 0.058666 (rank : 102) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VCM3, Q9D1E2 | Gene names | Zfyve21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 21. | |||||
|
ZFY21_HUMAN
|
||||||
| NC score | 0.057552 (rank : 103) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BQ24, Q86T05, Q96LT1 | Gene names | ZFYVE21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 21. | |||||
|
KR202_HUMAN
|
||||||
| NC score | 0.057146 (rank : 104) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3LI61 | Gene names | KRTAP20-2, KAP20.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 20-2. | |||||
|
ARHG9_HUMAN
|
||||||
| NC score | 0.056996 (rank : 105) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43307, Q5JSL6 | Gene names | ARHGEF9, KIAA0424 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 9 (Rac/Cdc42 guanine nucleotide exchange factor 9) (Collybistin) (PEM-2 homolog). | |||||
|
ARHG9_MOUSE
|
||||||
| NC score | 0.056533 (rank : 106) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3UTH8, Q3TQ60, Q80U06, Q8CAF9 | Gene names | Arhgef9, Kiaa0424 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 9 (Rac/Cdc42 guanine nucleotide exchange factor 9) (Collybistin). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.056119 (rank : 107) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.055435 (rank : 108) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
ZFY16_MOUSE
|
||||||
| NC score | 0.054869 (rank : 109) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80U44, Q8BRD2, Q8CG97 | Gene names | Zfyve16, Kiaa0305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 16 (Endofin) (Endosomal- associated FYVE domain protein). | |||||
|
SMUF2_HUMAN
|
||||||
| NC score | 0.054377 (rank : 110) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
SMUF1_HUMAN
|
||||||
| NC score | 0.054315 (rank : 111) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
SMUF1_MOUSE
|
||||||
| NC score | 0.054044 (rank : 112) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
ITCH_MOUSE
|
||||||
| NC score | 0.053536 (rank : 113) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
ITCH_HUMAN
|
||||||
| NC score | 0.053181 (rank : 114) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
ZFY16_HUMAN
|
||||||
| NC score | 0.053169 (rank : 115) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z3T8, O15023, Q7LAU7, Q86T69, Q8N5L3, Q8NEK3 | Gene names | ZFYVE16, KIAA0305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 16 (Endofin) (Endosome- associated FYVE domain protein). | |||||
|
WWP2_MOUSE
|
||||||
| NC score | 0.053052 (rank : 116) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
WWP2_HUMAN
|
||||||
| NC score | 0.052888 (rank : 117) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
GNRP_MOUSE
|
||||||
| NC score | 0.052682 (rank : 118) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
NGEF_HUMAN
|
||||||
| NC score | 0.052048 (rank : 119) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N5V2, Q53QQ4, Q53ST7, Q6GMQ5, Q9NQD6 | Gene names | NGEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephexin-1 (Eph-interacting exchange protein) (Neuronal guanine nucleotide exchange factor). | |||||
|
ARHG5_HUMAN
|
||||||
| NC score | 0.051662 (rank : 120) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12774 | Gene names | ARHGEF5, TIM | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 5 (Guanine nucleotide regulatory protein TIM) (Oncogene TIM) (p60 TIM) (Transforming immortalized mammary oncogene). | |||||
|
WWP1_MOUSE
|
||||||
| NC score | 0.050949 (rank : 121) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
WWP1_HUMAN
|
||||||
| NC score | 0.050586 (rank : 122) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
GNRP_HUMAN
|
||||||
| NC score | 0.050288 (rank : 123) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
NGEF_MOUSE
|
||||||
| NC score | 0.050128 (rank : 124) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHT1, Q8R204, Q923H2, Q9JHT9 | Gene names | Ngef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephexin-1 (Eph-interacting exchange protein) (Neuronal guanine nucleotide exchange factor). | |||||
|
MCF2_HUMAN
|
||||||
| NC score | 0.050082 (rank : 125) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P10911, P14919, Q5JYJ4, Q8IUF3, Q8IUF4, Q9UJB3 | Gene names | MCF2, DBL | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene DBL (Proto-oncogene MCF-2) [Contains: MCF2-transforming protein; DBL-transforming protein]. | |||||
|
OGT1_HUMAN
|
||||||
| NC score | 0.046099 (rank : 126) | θ value | 0.0961366 (rank : 69) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
TR11B_MOUSE
|
||||||
| NC score | 0.030243 (rank : 127) | θ value | 0.47712 (rank : 76) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08712, O70202 | Gene names | Tnfrsf11b, Ocif, Opg | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11B precursor (Osteoprotegerin) (Osteoclastogenesis inhibitory factor). | |||||
|
PLEK_HUMAN
|
||||||
| NC score | 0.028667 (rank : 128) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
SNX1_HUMAN
|
||||||
| NC score | 0.028557 (rank : 129) | θ value | 1.06291 (rank : 78) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13596, O60750, O60751 | Gene names | SNX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-1. | |||||
|
CENA1_HUMAN
|
||||||
| NC score | 0.025002 (rank : 130) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75689 | Gene names | CENTA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 1 (Putative MAPK-activating protein PM25). | |||||
|
PKHA1_HUMAN
|
||||||
| NC score | 0.024487 (rank : 131) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
POPD2_HUMAN
|
||||||
| NC score | 0.022614 (rank : 132) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HBU9, Q86UE7 | Gene names | POPDC2, POP2 | |||
|
Domain Architecture |

|
|||||
| Description | Popeye domain-containing protein 2 (Popeye protein 2). | |||||
|
PANK4_HUMAN
|
||||||
| NC score | 0.022441 (rank : 133) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NVE7, Q53EU3, Q5TA84, Q7RTX3, Q9H3X5 | Gene names | PANK4 | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 4 (EC 2.7.1.33) (Pantothenic acid kinase 4) (hPanK4). | |||||
|
CENA2_HUMAN
|
||||||
| NC score | 0.017934 (rank : 134) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NPF8, Q8N4Q6, Q96SD5 | Gene names | CENTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-alpha 2. | |||||
|
CEND1_HUMAN
|
||||||
| NC score | 0.017891 (rank : 135) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
HCFC1_MOUSE
|
||||||
| NC score | 0.017376 (rank : 136) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
HCFC1_HUMAN
|
||||||
| NC score | 0.017201 (rank : 137) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
LPIN2_HUMAN
|
||||||
| NC score | 0.016133 (rank : 138) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92539 | Gene names | LPIN2, KIAA0249 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-2. | |||||
|
LPIN2_MOUSE
|
||||||
| NC score | 0.015679 (rank : 139) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99PI5, Q8C357, Q8C7I8, Q8CC85, Q8CHR7 | Gene names | Lpin2 | |||
|
Domain Architecture |

|
|||||
| Description | Lipin-2. | |||||
|
CEND1_MOUSE
|
||||||
| NC score | 0.015470 (rank : 140) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
ATR_HUMAN
|
||||||
| NC score | 0.015016 (rank : 141) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13535, Q7KYL3, Q93051, Q9BXK4 | Gene names | ATR, FRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein) (FRAP-related protein 1). | |||||
|
VKGC_MOUSE
|
||||||
| NC score | 0.014577 (rank : 142) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QYC7, Q3UXN5, Q8CCB3 | Gene names | Ggcx | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent gamma-carboxylase (EC 6.4.-.-) (Gamma-glutamyl carboxylase) (Vitamin K gamma glutamyl carboxylase). | |||||
|
KLH14_HUMAN
|
||||||
| NC score | 0.014514 (rank : 143) | θ value | 0.62314 (rank : 77) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P2G3 | Gene names | KLHL14, KIAA1384 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 14. | |||||
|
VKGC_HUMAN
|
||||||
| NC score | 0.014476 (rank : 144) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38435, Q14415, Q6GU45 | Gene names | GGCX, GC | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent gamma-carboxylase (EC 6.4.-.-) (Gamma-glutamyl carboxylase) (Vitamin K gamma glutamyl carboxylase). | |||||
|
E2AK4_MOUSE
|
||||||
| NC score | 0.012509 (rank : 145) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 984 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QZ05, Q6GQT4, Q6ZPT5, Q8C5S0, Q8CIF5, Q9CT30, Q9CUV9, Q9ESB6, Q9ESB7, Q9ESB8 | Gene names | Eif2ak4, Gcn2, Kiaa1338 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 2-alpha kinase 4 (EC 2.7.11.1) (GCN2-like protein) (mGCN2). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.011995 (rank : 146) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.011837 (rank : 147) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
K0406_MOUSE
|
||||||
| NC score | 0.011444 (rank : 148) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91V83, Q9CSY0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0406 homolog. | |||||
|
PK3CD_MOUSE
|
||||||
| NC score | 0.011297 (rank : 149) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35904 | Gene names | Pik3cd | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
MARK2_MOUSE
|
||||||
| NC score | 0.010091 (rank : 150) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q05512, Q6PDR4, Q8BR95 | Gene names | Mark2, Emk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL Motif Kinase) (EMK1). | |||||
|
DKC1_HUMAN
|
||||||
| NC score | 0.009793 (rank : 151) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60832, O43845, Q96G67, Q9Y505 | Gene names | DKC1, NOLA4 | |||
|
Domain Architecture |

|
|||||
| Description | H/ACA ribonucleoprotein complex subunit 4 (EC 5.4.99.-) (Dyskerin) (Nucleolar protein family A member 4) (snoRNP protein DKC1) (Nopp140- associated protein of 57 kDa) (Nucleolar protein NAP57) (CBF5 homolog). | |||||
|
KLHL3_HUMAN
|
||||||
| NC score | 0.008962 (rank : 152) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UH77, Q9UH75, Q9UH76, Q9ULU0, Q9Y6V6 | Gene names | KLHL3, KIAA1129 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 3. | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.008550 (rank : 153) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.008245 (rank : 154) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
R113A_HUMAN
|
||||||
| NC score | 0.008244 (rank : 155) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15541 | Gene names | RNF113A, RNF113, ZNF183 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113A (Zinc finger protein 183). | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.008070 (rank : 156) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
PRAX_HUMAN
|
||||||
| NC score | 0.007359 (rank : 157) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
PDLI1_HUMAN
|
||||||
| NC score | 0.005910 (rank : 158) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00151, Q5VZH5, Q9BPZ9 | Gene names | PDLIM1, CLIM1, CLP36 | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 1 (Elfin) (LIM domain protein CLP-36) (C- terminal LIM domain protein 1). | |||||
|
CSMD2_HUMAN
|
||||||
| NC score | 0.005371 (rank : 159) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.005340 (rank : 160) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
CCNB3_HUMAN
|
||||||
| NC score | 0.003688 (rank : 161) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | -0.001744 (rank : 162) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 111 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||