Please be patient as the page loads
|
ZFY16_HUMAN
|
||||||
| SwissProt Accessions | Q7Z3T8, O15023, Q7LAU7, Q86T69, Q8N5L3, Q8NEK3 | Gene names | ZFYVE16, KIAA0305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 16 (Endofin) (Endosome- associated FYVE domain protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ZFY16_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | Q7Z3T8, O15023, Q7LAU7, Q86T69, Q8N5L3, Q8NEK3 | Gene names | ZFYVE16, KIAA0305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 16 (Endofin) (Endosome- associated FYVE domain protein). | |||||
|
ZFY16_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.983444 (rank : 2) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q80U44, Q8BRD2, Q8CG97 | Gene names | Zfyve16, Kiaa0305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 16 (Endofin) (Endosomal- associated FYVE domain protein). | |||||
|
ZFYV9_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.947491 (rank : 3) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O95405, Q9UNE1, Q9Y5R7 | Gene names | ZFYVE9, MADHIP, SARA, SMADIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 9 (Mothers against decapentaplegic homolog-interacting protein) (Madh-interacting protein) (Smad anchor for receptor activation) (Receptor activation anchor) (hSARA) (Novel serine protease) (NSP). | |||||
|
FGD6_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 4) | NC score | 0.545512 (rank : 12) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
FGD6_MOUSE
|
||||||
| θ value | 5.25075e-16 (rank : 5) | NC score | 0.533390 (rank : 16) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
ZFY21_MOUSE
|
||||||
| θ value | 4.91457e-14 (rank : 6) | NC score | 0.726606 (rank : 4) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8VCM3, Q9D1E2 | Gene names | Zfyve21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 21. | |||||
|
FGD5_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 7) | NC score | 0.539289 (rank : 14) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
ZFY21_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 8) | NC score | 0.718822 (rank : 5) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9BQ24, Q86T05, Q96LT1 | Gene names | ZFYVE21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 21. | |||||
|
HGS_MOUSE
|
||||||
| θ value | 5.43371e-13 (rank : 9) | NC score | 0.528832 (rank : 18) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
HGS_HUMAN
|
||||||
| θ value | 1.58096e-12 (rank : 10) | NC score | 0.529031 (rank : 17) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
FYV1_HUMAN
|
||||||
| θ value | 2.69671e-12 (rank : 11) | NC score | 0.509479 (rank : 19) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
FGD4_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 12) | NC score | 0.465681 (rank : 24) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96M96, Q6ULS2, Q8TCP6 | Gene names | FGD4, FRABP, ZFYVE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein) (Zinc finger FYVE domain-containing protein 6). | |||||
|
FGD5_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 13) | NC score | 0.547082 (rank : 11) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q80UZ0, Q8BHM5 | Gene names | Fgd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5. | |||||
|
FYV1_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 14) | NC score | 0.506885 (rank : 20) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z1T6 | Gene names | Pip5k3, Pikfyve | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (PIP5K) (PtdIns(4)P-5- kinase) (PIKfyve) (p235). | |||||
|
FYCO1_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 15) | NC score | 0.271554 (rank : 45) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
PKHF2_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 16) | NC score | 0.659119 (rank : 8) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H8W4 | Gene names | PLEKHF2, ZFYVE18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 2 (PH domain- containing family F member 2) (PH and FYVE domain-containing protein 2) (Phafin-2) (Zinc finger FYVE domain-containing protein 18). | |||||
|
PKHF2_MOUSE
|
||||||
| θ value | 7.84624e-12 (rank : 17) | NC score | 0.659670 (rank : 7) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q91WB4 | Gene names | Plekhf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 2 (PH domain- containing family F member 2). | |||||
|
FGD4_MOUSE
|
||||||
| θ value | 1.02475e-11 (rank : 18) | NC score | 0.461930 (rank : 26) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
FGD1_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 19) | NC score | 0.459735 (rank : 27) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
FGD2_MOUSE
|
||||||
| θ value | 2.28291e-11 (rank : 20) | NC score | 0.484468 (rank : 21) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BY35, O88841, Q7TSE3, Q8VDH4 | Gene names | Fgd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 2. | |||||
|
MTMR3_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 21) | NC score | 0.389122 (rank : 34) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q13615, Q9NYN5, Q9NYN6, Q9UDX6, Q9UEG3 | Gene names | MTMR3, KIAA0371, ZFYVE10 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48) (FYVE domain-containing dual specificity protein phosphatase 1) (FYVE-DSP1) (Zinc finger FYVE domain-containing protein 10). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 22) | NC score | 0.298917 (rank : 44) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
FGD1_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 23) | NC score | 0.458899 (rank : 28) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
FGD2_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 24) | NC score | 0.483962 (rank : 22) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7Z6J4, Q5T8I1, Q6P6A8, Q6ZNL5, Q8IZ32, Q8N868, Q9H7M2 | Gene names | FGD2, ZFYVE4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 2 (Zinc finger FYVE domain-containing protein 4). | |||||
|
FYCO1_MOUSE
|
||||||
| θ value | 8.67504e-11 (rank : 25) | NC score | 0.252002 (rank : 47) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
RUFY1_MOUSE
|
||||||
| θ value | 8.67504e-11 (rank : 26) | NC score | 0.378348 (rank : 36) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 27) | NC score | 0.226768 (rank : 49) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
RUFY1_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 28) | NC score | 0.370120 (rank : 40) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
RUFY2_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 29) | NC score | 0.373518 (rank : 38) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8R4C2, Q69ZH1 | Gene names | Rufy2, Kiaa1537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Leucine zipper FYVE-finger protein) (LZ-FYVE). | |||||
|
ZFY28_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 30) | NC score | 0.671720 (rank : 6) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9HCC9 | Gene names | ZFYVE28, KIAA1643 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 28. | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 31) | NC score | 0.209125 (rank : 50) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
WDFY3_MOUSE
|
||||||
| θ value | 1.25267e-09 (rank : 32) | NC score | 0.374237 (rank : 37) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6VNB8, Q8C8H7 | Gene names | Wdfy3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Beach domain, WD repeat and FYVE domain-containing protein 1) (BWF1). | |||||
|
WDFY3_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 33) | NC score | 0.372749 (rank : 39) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8IZQ1, Q4W5K5, Q6P0Q5, Q8N1T2, Q96BS7, Q96N85, Q9Y2J7 | Gene names | WDFY3, KIAA0993 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Autophagy-linked FYVE protein) (Alfy). | |||||
|
PKHF1_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 34) | NC score | 0.642571 (rank : 10) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q3TB82, Q99M16 | Gene names | Plekhf1, Lapf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains). | |||||
|
PKHF1_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 35) | NC score | 0.646248 (rank : 9) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96S99, Q96K11, Q9BUB9 | Gene names | PLEKHF1, APPD, LAPF, ZFYVE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (PH and FYVE domain-containing protein 1) (Phafin-1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains) (Apoptosis-inducing protein) (Zinc finger FYVE domain-containing protein 15). | |||||
|
ANFY1_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 36) | NC score | 0.252027 (rank : 46) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q810B6, O54807, Q80TG6, Q80UH8 | Gene names | Ankfy1, Ankhzn, Kiaa1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 37) | NC score | 0.106399 (rank : 67) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ZFYV1_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 38) | NC score | 0.540492 (rank : 13) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9HBF4, Q8WYX7, Q96K57, Q9BXP9, Q9HCI3 | Gene names | ZFYVE1, DFCP1, KIAA1589, TAFF1, ZNFN2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 1 (Double FYVE-containing protein 1) (Tandem FYVE fingers-1) (SR3). | |||||
|
ZFYV1_MOUSE
|
||||||
| θ value | 8.97725e-08 (rank : 39) | NC score | 0.536831 (rank : 15) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q810J8 | Gene names | Zfyve1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 1. | |||||
|
ANFY1_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 40) | NC score | 0.246892 (rank : 48) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9P2R3, Q9ULG5 | Gene names | ANKFY1, ANKHZN, KIAA1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
FGD3_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 41) | NC score | 0.422982 (rank : 31) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q5JSP0, Q7Z7D9, Q8N5G1 | Gene names | FGD3, ZFYVE5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3 (Zinc finger FYVE domain-containing protein 5). | |||||
|
WDFY2_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 42) | NC score | 0.361030 (rank : 43) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96P53, Q96CS1 | Gene names | WDFY2, WDF2, ZFYVE22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 2 (WD40- and FYVE domain- containing protein 2) (Zinc finger FYVE domain-containing protein 22). | |||||
|
WDFY2_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 43) | NC score | 0.363861 (rank : 42) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BUB4 | Gene names | Wdfy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 2 (WD40- and FYVE domain- containing protein 2). | |||||
|
RBNS5_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 44) | NC score | 0.468950 (rank : 23) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H1K0, Q3KP30, Q59EY8, Q8NAQ1 | Gene names | ZFYVE20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20) (110 kDa protein). | |||||
|
RBNS5_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 45) | NC score | 0.464873 (rank : 25) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q80Y56, Q8K0L6, Q9CTW0 | Gene names | Zfyve20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20). | |||||
|
FGD3_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 46) | NC score | 0.383885 (rank : 35) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O88842, Q8BQ72 | Gene names | Fgd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3. | |||||
|
WDFY1_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 47) | NC score | 0.369723 (rank : 41) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8IWB7, Q9H9D5, Q9P2B3 | Gene names | WDFY1, KIAA1435, WDF1, ZFYVE17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 1 (WD40- and FYVE domain- containing protein 1) (Phosphoinositide-binding protein 1) (FENS-1) (Zinc finger FYVE domain-containing protein 17). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 48) | NC score | 0.093249 (rank : 74) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ZFY27_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 49) | NC score | 0.393501 (rank : 33) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5T4F4, Q5T4F1, Q5T4F2, Q5T4F3, Q8N1K0, Q8N6D6, Q8NCA0, Q8NDE4, Q96M08 | Gene names | ZFYVE27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 27. | |||||
|
ZFY19_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 50) | NC score | 0.458555 (rank : 29) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96K21, Q86WC2, Q8WU96 | Gene names | ZFYVE19, MPFYVE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 19 (MLL partner containing FYVE domain). | |||||
|
ZFY19_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 51) | NC score | 0.451573 (rank : 30) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9DAZ9, Q8VCV7 | Gene names | Zfyve19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 19. | |||||
|
ZFY27_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 52) | NC score | 0.399661 (rank : 32) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q3TXX3, Q8CFP8 | Gene names | Zfyve27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 27. | |||||
|
RFFL_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 53) | NC score | 0.187972 (rank : 51) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6ZQM0, Q5SVC2, Q5SVC4, Q9D543, Q9D9B1 | Gene names | Rffl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rififylin (RING finger and FYVE-like domain-containing protein 1) (Fring). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 54) | NC score | 0.031062 (rank : 133) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
RNF34_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 55) | NC score | 0.177297 (rank : 53) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q969K3, Q8NG47, Q9H6W8 | Gene names | RNF34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 34 (RING finger protein RIFF) (FYVE-RING finger protein Momo) (Human RING finger homologous to inhibitor of apoptosis protein) (hRFI) (Caspases-8 and -10-associated RING finger protein 1) (CARP-1) (Caspase regulator CARP1). | |||||
|
RNF34_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 56) | NC score | 0.177785 (rank : 52) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99KR6, Q3UV45 | Gene names | Rnf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 34 (RING finger protein RIFF) (Phafin-1). | |||||
|
RFFL_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 57) | NC score | 0.174657 (rank : 54) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WZ73, Q8NHW0, Q8TBY7, Q96BE6 | Gene names | RFFL, RNF189, RNF34L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rififylin (RING finger and FYVE-like domain-containing protein 1) (FYVE-RING finger protein Sakura) (Fring) (Caspases-8 and -10- associated RING finger protein 2) (CARP-2) (Caspase regulator CARP2) (RING finger protein 189) (RING finger protein 34-like). | |||||
|
WDR37_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 58) | NC score | 0.043672 (rank : 117) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2I8, Q5SW03, Q9NTJ6 | Gene names | WDR37, KIAA0982 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 37. | |||||
|
WDR37_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 59) | NC score | 0.042458 (rank : 118) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CBE3, Q80Y96, Q8CCL2 | Gene names | Wdr37, Kiaa0982 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 37. | |||||
|
BIN1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 60) | NC score | 0.032009 (rank : 132) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08539, Q62434 | Gene names | Bin1, Amphl, Sh3p9 | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (SH3-domain-containing protein 9). | |||||
|
AIM1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 61) | NC score | 0.037478 (rank : 121) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4K1, O00296, Q5VWJ2 | Gene names | AIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Absent in melanoma 1 protein. | |||||
|
GFRA2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 62) | NC score | 0.032807 (rank : 131) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08842 | Gene names | Gfra2, Gdnfrb, Trnr2 | |||
|
Domain Architecture |
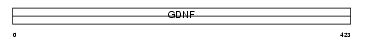
|
|||||
| Description | GDNF family receptor alpha-2 precursor (GFR-alpha-2) (Neurturin receptor alpha) (NTNR-alpha) (NRTNR-alpha) (TGF-beta-related neurotrophic factor receptor 2) (GDNF receptor beta) (GDNFR-beta). | |||||
|
ZN295_HUMAN
|
||||||
| θ value | 0.21417 (rank : 63) | NC score | 0.011464 (rank : 157) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULJ3, Q6P4R0 | Gene names | ZNF295, KIAA1227, ZBTB21 | |||
|
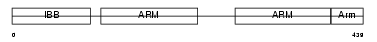
Domain Architecture |
|
|||||
| Description | Zinc finger protein 295 (Zinc finger and BTB domain-containing protein 21). | |||||
|
CP4AC_MOUSE
|
||||||
| θ value | 0.365318 (rank : 64) | NC score | 0.008898 (rank : 161) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91WL5, Q3UNE4, Q6P931, Q8N7N3 | Gene names | Cyp4a12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 4A12 (EC 1.14.14.1) (CYPIVA12). | |||||
|
IMA5_HUMAN
|
||||||
| θ value | 0.365318 (rank : 65) | NC score | 0.036438 (rank : 122) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15131 | Gene names | KPNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-6 subunit (Karyopherin alpha-5 subunit). | |||||
|
WEE1B_HUMAN
|
||||||
| θ value | 0.365318 (rank : 66) | NC score | 0.007057 (rank : 165) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P0C1S8 | Gene names | WEE1B, WEE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wee1-like protein kinase 1B (EC 2.7.10.2) (Wee1B kinase). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 0.47712 (rank : 67) | NC score | 0.012086 (rank : 154) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
IMA1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 68) | NC score | 0.035505 (rank : 123) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P52294, Q9BQ56 | Gene names | KPNA1, RCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (NPI-1). | |||||
|
SP2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 69) | NC score | 0.005567 (rank : 169) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02086 | Gene names | SP2, KIAA0048 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp2. | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 1.06291 (rank : 70) | NC score | 0.037518 (rank : 120) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
SCAR3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 71) | NC score | 0.017501 (rank : 148) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6AZY7, Q9UM15, Q9UM16 | Gene names | SCARA3, CSR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class A member 3 (Cellular stress response gene protein). | |||||
|
ZN230_HUMAN
|
||||||
| θ value | 1.06291 (rank : 72) | NC score | 0.001034 (rank : 179) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UIE0, O15322, Q9P1U6 | Gene names | ZNF230, FDZF2 | |||
|
Domain Architecture |
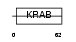
|
|||||
| Description | Zinc finger protein 230 (Zinc finger protein FDZF2). | |||||
|
CC14A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.016739 (rank : 150) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UNH5, O43171, O60727, O60728, Q8IXX0 | Gene names | CDC14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
IMA1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.034338 (rank : 126) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60960, Q3TF32 | Gene names | Kpna1, Rch2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (Importin alpha S1). | |||||
|
IMA7_MOUSE
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.033854 (rank : 128) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35345, Q9CVP9 | Gene names | Kpna6, Kpna5 | |||
|
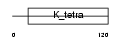
Domain Architecture |
|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6 subunit) (Importin alpha S2). | |||||
|
PODXL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.023732 (rank : 137) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00592 | Gene names | PODXL, PCLP, PCLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
RFIP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.018946 (rank : 146) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D620, Q3UBC2, Q8BN24 | Gene names | Rab11fip1, Rcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 78) | NC score | 0.035018 (rank : 125) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
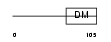
DMRT1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.021523 (rank : 141) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZ59, Q9QZ37, Q9QZA4 | Gene names | Dmrt1 | |||
|
Domain Architecture |
|
|||||
| Description | Doublesex- and mab-3-related transcription factor 1. | |||||
|
FINC_MOUSE
|
||||||
| θ value | 1.81305 (rank : 80) | NC score | 0.007041 (rank : 166) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.037744 (rank : 119) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
IMA7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 82) | NC score | 0.033264 (rank : 130) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60684 | Gene names | KPNA6, IPOA7 | |||
|
Domain Architecture |
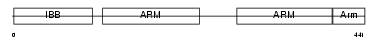
|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6). | |||||
|
NEB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 83) | NC score | 0.035261 (rank : 124) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
SAFB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 84) | NC score | 0.016166 (rank : 151) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15424, O60406, Q59HH8 | Gene names | SAFB, HAP, HET, SAFB1 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B (Scaffold attachment factor B1) (SAF-B) (HSP27 estrogen response element-TATA box-binding protein) (HSP27 ERE- TATA-binding protein). | |||||
|
SAS10_HUMAN
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.023112 (rank : 138) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQZ2, Q6FI82 | Gene names | CRLZ1, SAS10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Something about silencing protein 10 (Disrupter of silencing SAS10) (Charged amino acid-rich leucine zipper 1). | |||||
|
ZN408_HUMAN
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.001415 (rank : 177) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H9D4 | Gene names | ZNF408, PFM14, PRDM17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 408 (PR-domain zinc finger protein 17). | |||||
|
AFTIN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.033877 (rank : 127) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80WT5, Q5SSE6, Q99KJ1 | Gene names | Aftph, Afth | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aftiphilin. | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.020558 (rank : 143) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
MUC13_HUMAN
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | 0.021589 (rank : 140) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3R2, Q6UWD9, Q9NXT5 | Gene names | MUC13, DRCC1, RECC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-13 precursor (Down-regulated in colon cancer 1). | |||||
|
NU153_HUMAN
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.030215 (rank : 134) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
WEE1B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.004674 (rank : 171) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q66JT0, Q4U4S4, Q7TPV9 | Gene names | Wee1b, Wee2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wee1-like protein kinase 1B (EC 2.7.10.2) (Wee1B kinase) (mWee1B). | |||||
|
DGKG_HUMAN
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.010160 (rank : 158) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49619 | Gene names | DGKG, DAGK3 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase gamma (EC 2.7.1.107) (Diglyceride kinase gamma) (DGK-gamma) (DAG kinase gamma). | |||||
|
GFRA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.023930 (rank : 136) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00451, O15316, O15328, Q7Z5C2 | Gene names | GFRA2, GDNFRB, RETL2, TRNR2 | |||
|
Domain Architecture |
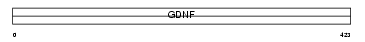
|
|||||
| Description | GDNF family receptor alpha-2 precursor (GFR-alpha-2) (Neurturin receptor alpha) (NTNR-alpha) (NRTNR-alpha) (TGF-beta-related neurotrophic factor receptor 2) (GDNF receptor beta) (GDNFR-beta) (RET ligand 2). | |||||
|
ZN317_HUMAN
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.000869 (rank : 183) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96PQ6, Q96PM0, Q96PM1, Q96PT2 | Gene names | ZNF317 | |||
|
Domain Architecture |
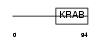
|
|||||
| Description | Zinc finger protein 317. | |||||
|
CND2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.022077 (rank : 139) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15003, Q8TB87 | Gene names | NCAPH, BRRN, BRRN1, CAPH, KIAA0074 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 2 (Non-SMC condensin I complex subunit H) (Barren homolog protein 1) (Chromosome-associated protein H) (hCAP-H) (XCAP-H homolog). | |||||
|
MAGBI_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.005141 (rank : 170) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96M61 | Gene names | MAGEB18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen B18 (MAGE-B18 antigen). | |||||
|
PCD12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.000995 (rank : 180) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NPG4, Q6UXB6, Q96KB8, Q9H7Y6, Q9H8E0 | Gene names | PCDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-12 precursor (Vascular cadherin-2) (Vascular endothelial cadherin-2) (VE-cadherin-2) (VE-cad-2). | |||||
|
ZAN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.006147 (rank : 168) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ZN555_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | -0.000154 (rank : 184) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NEP9, Q8NA46, Q96MP1 | Gene names | ZNF555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 555. | |||||
|
ZN699_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.000932 (rank : 182) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q32M78, Q8N9A1 | Gene names | ZNF699 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 699 (Hangover homolog). | |||||
|
ZO3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.006627 (rank : 167) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95049 | Gene names | TJP3, ZO3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
CEP57_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.033514 (rank : 129) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 580 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CEE0, Q6ZQJ3, Q7TN18, Q80X65, Q810F2, Q9D4J4, Q9D5S4, Q9D5W5 | Gene names | Cep57, Kiaa0092, Tsp57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin). | |||||
|
CND2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.021074 (rank : 142) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C156, Q8BY01, Q8VDM9 | Gene names | Ncaph, Brrn, Brrn1, Caph | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 2 (Non-SMC condensin I complex subunit H) (Barren homolog protein 1) (Chromosome-associated protein H) (mCAP-H) (XCAP-H homolog). | |||||
|
DCR1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.011607 (rank : 155) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PJP8, Q14701, Q6P5Y3, Q6PKL4 | Gene names | DCLRE1A, KIAA0086, SNM1, SNM1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA cross-link repair 1A protein (hSNM1) (hSNM1A). | |||||
|
DGKG_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.008839 (rank : 162) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WG7 | Gene names | Dgkg, Dagk3 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase gamma (EC 2.7.1.107) (Diglyceride kinase gamma) (DGK-gamma) (DAG kinase gamma) (88 kDa diacylglycerol kinase). | |||||
|
DLG7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.012374 (rank : 153) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15398, Q8NG58 | Gene names | DLG7, KIAA0008 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 7 (Hepatoma up-regulated protein) (HURP). | |||||
|
EAF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.013667 (rank : 152) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96CJ1, Q9NZ82 | Gene names | EAF2, TRAITS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELL-associated factor 2 (Testosterone-regulated apoptosis inducer and tumor suppressor protein). | |||||
|
K2027_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.019032 (rank : 145) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZVF9, Q8IVE4 | Gene names | KIAA2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
NR1H2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.002197 (rank : 173) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60644 | Gene names | Nr1h2, Lxrb, Rip15, Unr, Unr2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterols receptor LXR-beta (Liver X receptor beta) (Nuclear orphan receptor LXR-beta) (Ubiquitously-expressed nuclear receptor) (Retinoid X receptor-interacting protein No.15). | |||||
|
T2FA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.019714 (rank : 144) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
AFAD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.008995 (rank : 160) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
CNTP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.003569 (rank : 172) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZ76, Q9C0E9 | Gene names | CNTNAP3, CASPR3, KIAA1714 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 3 precursor (Cell recognition molecule Caspr3). | |||||
|
CUBN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | -0.000156 (rank : 185) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60494, Q5VTA6, Q96RU9 | Gene names | CUBN, IFCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor) (Intrinsic factor-vitamin B12 receptor) (460 kDa receptor) (Intestinal intrinsic factor receptor). | |||||
|
PO2F1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.001854 (rank : 174) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14859, Q5TBT7, Q6PK46, Q8NEU9, Q9BPV1 | Gene names | POU2F1, OCT1, OTF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 1 (Octamer-binding transcription factor 1) (Oct-1) (OTF-1) (NF-A1). | |||||
|
RBP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.007807 (rank : 163) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ERU9, Q61992, Q8C9K9 | Gene names | Ranbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2 (RanBP2). | |||||
|
TRI37_MOUSE
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.007467 (rank : 164) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PCX9, Q8CHC5 | Gene names | Trim37, Kiaa0898 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 37. | |||||
|
ZFY1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.001067 (rank : 178) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10925 | Gene names | Zfy1, Zfy-1 | |||
|
Domain Architecture |
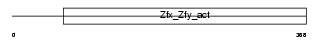
|
|||||
| Description | Zinc finger Y-chromosomal protein 1. | |||||
|
ARID2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.011502 (rank : 156) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
MAZ_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.001470 (rank : 175) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P56270, Q15703, Q99443 | Gene names | MAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myc-associated zinc finger protein (MAZI) (Purine-binding transcription factor) (Pur-1) (ZF87) (ZIF87). | |||||
|
MAZ_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.001432 (rank : 176) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P56671, Q9R1W0 | Gene names | Maz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myc-associated zinc finger protein (MAZI) (Purine-binding transcription factor) (Pur-1). | |||||
|
SDPR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.026276 (rank : 135) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95810 | Gene names | SDPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68). | |||||
|
SMYD5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.009089 (rank : 159) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3TYX3, Q3TRB2, Q91YL6 | Gene names | Smyd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 5 (Protein NN8-4AG). | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.017230 (rank : 149) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.018639 (rank : 147) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
VEZF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.000980 (rank : 181) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14119 | Gene names | VEZF1, DB1, ZNF161 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vascular endothelial zinc finger 1 (Zinc finger protein 161) (Putative transcription factor DB1). | |||||
|
ABR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.082540 (rank : 85) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
ALS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.053169 (rank : 109) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
|
ARHG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.082165 (rank : 86) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92888, O00513, Q8N4J4, Q96BF4, Q96F17, Q9BSB1 | Gene names | ARHGEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (p115-RhoGEF) (p115RhoGEF) (115 kDa guanine nucleotide exchange factor) (Sub1.5). | |||||
|
ARHG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.081413 (rank : 87) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
ARHG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.068471 (rank : 91) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92974, O75142, Q15079 | Gene names | ARHGEF2, KIAA0651, LFP40 | |||
|
Domain Architecture |
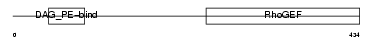
|
|||||
| Description | Rho/Rac guanine nucleotide exchange factor 2 (GEF-H1 protein) (Proliferating cell nucleolar antigen p40). | |||||
|
ARHG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.093870 (rank : 73) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60875, O09115 | Gene names | Arhgef2, Lbcl1, Lfc | |||
|
Domain Architecture |
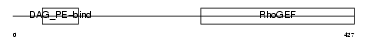
|
|||||
| Description | Rho/Rac guanine nucleotide exchange factor 2 (Lymphoid blast crisis- like 1) (LBC'S first cousin) (Oncogene LFC) (RHOBIN). | |||||
|
ARHG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.052788 (rank : 112) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NR81, Q6NUN3, Q7Z4U2, Q7Z5T2, Q9H7T4 | Gene names | ARHGEF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 3 (Exchange factor found in platelets and leukemic and neuronal tissues) (XPLN). | |||||
|
ARHG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.054645 (rank : 106) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91X46, Q8CDM0, Q91VY4, Q99K14, Q9DC31 | Gene names | Arhgef3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 3. | |||||
|
ARHG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.084947 (rank : 80) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NR80, Q9HDC6, Q9UPP0 | Gene names | ARHGEF4, KIAA1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
ARHG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.084254 (rank : 81) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TNR9, Q80TJ6 | Gene names | Arhgef4, Kiaa1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
ARHG5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.115256 (rank : 64) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q12774 | Gene names | ARHGEF5, TIM | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 5 (Guanine nucleotide regulatory protein TIM) (Oncogene TIM) (p60 TIM) (Transforming immortalized mammary oncogene). | |||||
|
ARHG7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.063408 (rank : 101) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14155, Q6P9G3, Q6PII2, Q86W63, Q8N3M1 | Gene names | ARHGEF7, COOL1, KIAA0142, P85SPR, PAK3BP, PIXB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (COOL-1) (p85). | |||||
|
ARHG7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.052648 (rank : 113) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ES28, O08757, Q9ES27 | Gene names | Arhgef7, Pak3bp | |||
|
Domain Architecture |
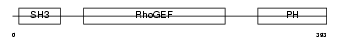
|
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (p85SPR). | |||||
|
ARHG8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.053050 (rank : 110) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z628, Q12773, Q96D82, Q99903, Q9UEN6 | Gene names | NET1, ARHGEF8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroepithelial cell-transforming gene 1 protein (p65 Net1 proto- oncogene) (Rho guanine nucleotide exchange factor 8). | |||||
|
ARHG9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.079060 (rank : 88) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43307, Q5JSL6 | Gene names | ARHGEF9, KIAA0424 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 9 (Rac/Cdc42 guanine nucleotide exchange factor 9) (Collybistin) (PEM-2 homolog). | |||||
|
ARHG9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.077734 (rank : 90) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3UTH8, Q3TQ60, Q80U06, Q8CAF9 | Gene names | Arhgef9, Kiaa0424 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 9 (Rac/Cdc42 guanine nucleotide exchange factor 9) (Collybistin). | |||||
|
ARHGA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.108949 (rank : 66) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15013, O14665, Q68D55, Q8IWD9, Q8IY77 | Gene names | ARHGEF10, KIAA0294 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 10. | |||||
|
ARHGA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.109695 (rank : 65) | |||
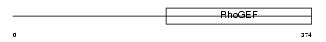
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C033, Q5DU38, Q80VH8, Q8BW76, Q922S7 | Gene names | Arhgef10, Kiaa0294 | |||
|
Domain Architecture |
|
|||||
| Description | Rho guanine nucleotide exchange factor 10. | |||||
|
ARHGB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.065621 (rank : 96) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
ARHGC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.064810 (rank : 98) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
ARHGC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.064012 (rank : 99) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R4H2, Q80U18 | Gene names | Arhgef12, Kiaa0382, Larg | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
ARHGF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.100205 (rank : 70) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O94989, Q8N449, Q9H8B4 | Gene names | ARHGEF15, KIAA0915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 15 (Vsm-RhoGEF). | |||||
|
ARHGG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.099910 (rank : 71) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5VV41, Q86TF0, Q99434 | Gene names | ARHGEF16, NBR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
ARHGG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.092554 (rank : 75) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3U5C8, Q501M8, Q8VCE8 | Gene names | Arhgef16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
BCR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.084035 (rank : 83) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
CJ064_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.078945 (rank : 89) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZS81, Q8N4A3 | Gene names | C10orf64 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf64. | |||||
|
ECT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.129160 (rank : 60) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H8V3, Q9NSV8, Q9NVW9 | Gene names | ECT2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
ECT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.132545 (rank : 59) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q07139 | Gene names | Ect2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
FARP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.136383 (rank : 58) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
FARP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.140941 (rank : 55) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
FARP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.139686 (rank : 56) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
GNRP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.084251 (rank : 82) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
GNRP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.083740 (rank : 84) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
HAPIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.105517 (rank : 68) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60229 | Gene names | HAPIP, DUO | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein-interacting protein (Protein Duo). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.067752 (rank : 93) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.067815 (rank : 92) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
ITSN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.067523 (rank : 94) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
ITSN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.067498 (rank : 95) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
LBC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.086057 (rank : 79) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12802 | Gene names | LBC | |||
|
Domain Architecture |
|
|||||
| Description | LBC oncogene (P47) (Lymphoid blast crisis oncogene). | |||||
|
MCF2L_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.091079 (rank : 76) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q64096 | Gene names | Mcf2l, Dbs | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide exchange factor DBS (DBL's big sister) (MCF2- transforming sequence-like protein). | |||||
|
MCF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.090044 (rank : 77) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10911, P14919, Q5JYJ4, Q8IUF3, Q8IUF4, Q9UJB3 | Gene names | MCF2, DBL | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene DBL (Proto-oncogene MCF-2) [Contains: MCF2-transforming protein; DBL-transforming protein]. | |||||
|
NBEAL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.055177 (rank : 105) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZS30, Q6Y876, Q6ZQY5, Q96Q30 | Gene names | NBEAL1, ALS2CR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurobeachin-like 1 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 17 protein). | |||||
|
NGEF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.102749 (rank : 69) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N5V2, Q53QQ4, Q53ST7, Q6GMQ5, Q9NQD6 | Gene names | NGEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephexin-1 (Eph-interacting exchange protein) (Neuronal guanine nucleotide exchange factor). | |||||
|
NGEF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.099775 (rank : 72) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CHT1, Q8R204, Q923H2, Q9JHT9 | Gene names | Ngef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephexin-1 (Eph-interacting exchange protein) (Neuronal guanine nucleotide exchange factor). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.063277 (rank : 102) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.060560 (rank : 103) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.115853 (rank : 63) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ULL1, Q5T1F2 | Gene names | PLEKHG1, KIAA1209 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family G member 1. | |||||
|
PKHG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.088461 (rank : 78) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q58EX7, Q4G0J8, Q4H485, Q56A69, Q9H7K4, Q9UFW0 | Gene names | PLEKHG4, PRTPHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Puratrophin-1 (Pleckstrin homology domain-containing family G member 4) (Purkinje cell atrophy-associated protein 1). | |||||
|
PREX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.138099 (rank : 57) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TCU6, Q5JS95, Q5JS96, Q69YL2, Q7Z2L9, Q9BQH0, Q9BX55, Q9H4Q6, Q9P2D2, Q9UGQ4 | Gene names | PREX1, KIAA1415 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (P-Rex1 protein). | |||||
|
RUFY3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.116360 (rank : 62) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7L099, O94948, Q9UI00 | Gene names | RUFY3, KIAA0871 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RUFY3 (Rap2-interacting protein x) (RIPx). | |||||
|
RUFY3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.117456 (rank : 61) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 579 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D394, Q3U348, Q3UF96, Q6PE64, Q8VD10 | Gene names | Rufy3, D5Bwg0860e, Ripx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RUFY3 (Rap2-interacting protein x) (RIPx). | |||||
|
SWP70_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.053265 (rank : 108) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
SWP70_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.050740 (rank : 116) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
TIAM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.065588 (rank : 97) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13009 | Gene names | TIAM1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
TIAM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.063631 (rank : 100) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60610 | Gene names | Tiam1, Tiam-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
VAV2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.053040 (rank : 111) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60992 | Gene names | Vav2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
VAV3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.051964 (rank : 114) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKW4, O95230, Q9Y5X8 | Gene names | VAV3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
VAV3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.051291 (rank : 115) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9R0C8, Q7TS85 | Gene names | Vav3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
VAV_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.056854 (rank : 104) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P15498, Q15860 | Gene names | VAV1, VAV | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene vav. | |||||
|
VAV_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.054320 (rank : 107) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P27870, Q8BTV7 | Gene names | Vav1, Vav | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene vav (p95vav). | |||||
|
ZFY16_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | Q7Z3T8, O15023, Q7LAU7, Q86T69, Q8N5L3, Q8NEK3 | Gene names | ZFYVE16, KIAA0305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 16 (Endofin) (Endosome- associated FYVE domain protein). | |||||
|
ZFY16_MOUSE
|
||||||
| NC score | 0.983444 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q80U44, Q8BRD2, Q8CG97 | Gene names | Zfyve16, Kiaa0305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 16 (Endofin) (Endosomal- associated FYVE domain protein). | |||||
|
ZFYV9_HUMAN
|
||||||
| NC score | 0.947491 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O95405, Q9UNE1, Q9Y5R7 | Gene names | ZFYVE9, MADHIP, SARA, SMADIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 9 (Mothers against decapentaplegic homolog-interacting protein) (Madh-interacting protein) (Smad anchor for receptor activation) (Receptor activation anchor) (hSARA) (Novel serine protease) (NSP). | |||||
|
ZFY21_MOUSE
|
||||||
| NC score | 0.726606 (rank : 4) | θ value | 4.91457e-14 (rank : 6) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8VCM3, Q9D1E2 | Gene names | Zfyve21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 21. | |||||
|
ZFY21_HUMAN
|
||||||
| NC score | 0.718822 (rank : 5) | θ value | 2.43908e-13 (rank : 8) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9BQ24, Q86T05, Q96LT1 | Gene names | ZFYVE21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 21. | |||||
|
ZFY28_HUMAN
|
||||||
| NC score | 0.671720 (rank : 6) | θ value | 2.52405e-10 (rank : 30) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9HCC9 | Gene names | ZFYVE28, KIAA1643 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 28. | |||||
|
PKHF2_MOUSE
|
||||||
| NC score | 0.659670 (rank : 7) | θ value | 7.84624e-12 (rank : 17) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q91WB4 | Gene names | Plekhf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 2 (PH domain- containing family F member 2). | |||||
|
PKHF2_HUMAN
|
||||||
| NC score | 0.659119 (rank : 8) | θ value | 7.84624e-12 (rank : 16) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H8W4 | Gene names | PLEKHF2, ZFYVE18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 2 (PH domain- containing family F member 2) (PH and FYVE domain-containing protein 2) (Phafin-2) (Zinc finger FYVE domain-containing protein 18). | |||||
|
PKHF1_HUMAN
|
||||||
| NC score | 0.646248 (rank : 9) | θ value | 2.79066e-09 (rank : 35) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96S99, Q96K11, Q9BUB9 | Gene names | PLEKHF1, APPD, LAPF, ZFYVE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (PH and FYVE domain-containing protein 1) (Phafin-1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains) (Apoptosis-inducing protein) (Zinc finger FYVE domain-containing protein 15). | |||||
|
PKHF1_MOUSE
|
||||||
| NC score | 0.642571 (rank : 10) | θ value | 2.13673e-09 (rank : 34) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q3TB82, Q99M16 | Gene names | Plekhf1, Lapf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains). | |||||
|
FGD5_MOUSE
|
||||||
| NC score | 0.547082 (rank : 11) | θ value | 3.52202e-12 (rank : 13) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q80UZ0, Q8BHM5 | Gene names | Fgd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5. | |||||
|
FGD6_HUMAN
|
||||||
| NC score | 0.545512 (rank : 12) | θ value | 1.38178e-16 (rank : 4) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
ZFYV1_HUMAN
|
||||||
| NC score | 0.540492 (rank : 13) | θ value | 6.87365e-08 (rank : 38) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9HBF4, Q8WYX7, Q96K57, Q9BXP9, Q9HCI3 | Gene names | ZFYVE1, DFCP1, KIAA1589, TAFF1, ZNFN2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 1 (Double FYVE-containing protein 1) (Tandem FYVE fingers-1) (SR3). | |||||
|
FGD5_HUMAN
|
||||||
| NC score | 0.539289 (rank : 14) | θ value | 2.43908e-13 (rank : 7) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
ZFYV1_MOUSE
|
||||||
| NC score | 0.536831 (rank : 15) | θ value | 8.97725e-08 (rank : 39) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q810J8 | Gene names | Zfyve1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 1. | |||||
|
FGD6_MOUSE
|
||||||
| NC score | 0.533390 (rank : 16) | θ value | 5.25075e-16 (rank : 5) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
HGS_HUMAN
|
||||||
| NC score | 0.529031 (rank : 17) | θ value | 1.58096e-12 (rank : 10) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.528832 (rank : 18) | θ value | 5.43371e-13 (rank : 9) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
FYV1_HUMAN
|
||||||
| NC score | 0.509479 (rank : 19) | θ value | 2.69671e-12 (rank : 11) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
FYV1_MOUSE
|
||||||
| NC score | 0.506885 (rank : 20) | θ value | 3.52202e-12 (rank : 14) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z1T6 | Gene names | Pip5k3, Pikfyve | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (PIP5K) (PtdIns(4)P-5- kinase) (PIKfyve) (p235). | |||||
|
FGD2_MOUSE
|
||||||
| NC score | 0.484468 (rank : 21) | θ value | 2.28291e-11 (rank : 20) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BY35, O88841, Q7TSE3, Q8VDH4 | Gene names | Fgd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 2. | |||||
|
FGD2_HUMAN
|
||||||
| NC score | 0.483962 (rank : 22) | θ value | 5.08577e-11 (rank : 24) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7Z6J4, Q5T8I1, Q6P6A8, Q6ZNL5, Q8IZ32, Q8N868, Q9H7M2 | Gene names | FGD2, ZFYVE4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 2 (Zinc finger FYVE domain-containing protein 4). | |||||
|
RBNS5_HUMAN
|
||||||
| NC score | 0.468950 (rank : 23) | θ value | 4.92598e-06 (rank : 44) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H1K0, Q3KP30, Q59EY8, Q8NAQ1 | Gene names | ZFYVE20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20) (110 kDa protein). | |||||
|
FGD4_HUMAN
|
||||||
| NC score | 0.465681 (rank : 24) | θ value | 3.52202e-12 (rank : 12) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96M96, Q6ULS2, Q8TCP6 | Gene names | FGD4, FRABP, ZFYVE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein) (Zinc finger FYVE domain-containing protein 6). | |||||
|
RBNS5_MOUSE
|
||||||
| NC score | 0.464873 (rank : 25) | θ value | 4.92598e-06 (rank : 45) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q80Y56, Q8K0L6, Q9CTW0 | Gene names | Zfyve20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20). | |||||
|
FGD4_MOUSE
|
||||||
| NC score | 0.461930 (rank : 26) | θ value | 1.02475e-11 (rank : 18) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
FGD1_MOUSE
|
||||||
| NC score | 0.459735 (rank : 27) | θ value | 1.74796e-11 (rank : 19) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
FGD1_HUMAN
|
||||||
| NC score | 0.458899 (rank : 28) | θ value | 5.08577e-11 (rank : 23) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
ZFY19_HUMAN
|
||||||
| NC score | 0.458555 (rank : 29) | θ value | 0.00035302 (rank : 50) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96K21, Q86WC2, Q8WU96 | Gene names | ZFYVE19, MPFYVE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 19 (MLL partner containing FYVE domain). | |||||
|
ZFY19_MOUSE
|
||||||
| NC score | 0.451573 (rank : 30) | θ value | 0.00035302 (rank : 51) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9DAZ9, Q8VCV7 | Gene names | Zfyve19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 19. | |||||
|
FGD3_HUMAN
|
||||||
| NC score | 0.422982 (rank : 31) | θ value | 5.81887e-07 (rank : 41) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q5JSP0, Q7Z7D9, Q8N5G1 | Gene names | FGD3, ZFYVE5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3 (Zinc finger FYVE domain-containing protein 5). | |||||
|
ZFY27_MOUSE
|
||||||
| NC score | 0.399661 (rank : 32) | θ value | 0.00035302 (rank : 52) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q3TXX3, Q8CFP8 | Gene names | Zfyve27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 27. | |||||
|
ZFY27_HUMAN
|
||||||
| NC score | 0.393501 (rank : 33) | θ value | 0.000270298 (rank : 49) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5T4F4, Q5T4F1, Q5T4F2, Q5T4F3, Q8N1K0, Q8N6D6, Q8NCA0, Q8NDE4, Q96M08 | Gene names | ZFYVE27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 27. | |||||
|
MTMR3_HUMAN
|
||||||
| NC score | 0.389122 (rank : 34) | θ value | 3.89403e-11 (rank : 21) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q13615, Q9NYN5, Q9NYN6, Q9UDX6, Q9UEG3 | Gene names | MTMR3, KIAA0371, ZFYVE10 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48) (FYVE domain-containing dual specificity protein phosphatase 1) (FYVE-DSP1) (Zinc finger FYVE domain-containing protein 10). | |||||
|
FGD3_MOUSE
|
||||||
| NC score | 0.383885 (rank : 35) | θ value | 1.43324e-05 (rank : 46) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O88842, Q8BQ72 | Gene names | Fgd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3. | |||||
|
RUFY1_MOUSE
|
||||||
| NC score | 0.378348 (rank : 36) | θ value | 8.67504e-11 (rank : 26) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
WDFY3_MOUSE
|
||||||
| NC score | 0.374237 (rank : 37) | θ value | 1.25267e-09 (rank : 32) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6VNB8, Q8C8H7 | Gene names | Wdfy3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Beach domain, WD repeat and FYVE domain-containing protein 1) (BWF1). | |||||
|
RUFY2_MOUSE
|
||||||
| NC score | 0.373518 (rank : 38) | θ value | 1.9326e-10 (rank : 29) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8R4C2, Q69ZH1 | Gene names | Rufy2, Kiaa1537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Leucine zipper FYVE-finger protein) (LZ-FYVE). | |||||
|
WDFY3_HUMAN
|
||||||
| NC score | 0.372749 (rank : 39) | θ value | 1.63604e-09 (rank : 33) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8IZQ1, Q4W5K5, Q6P0Q5, Q8N1T2, Q96BS7, Q96N85, Q9Y2J7 | Gene names | WDFY3, KIAA0993 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Autophagy-linked FYVE protein) (Alfy). | |||||
|
RUFY1_HUMAN
|
||||||
| NC score | 0.370120 (rank : 40) | θ value | 1.9326e-10 (rank : 28) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
WDFY1_HUMAN
|
||||||
| NC score | 0.369723 (rank : 41) | θ value | 1.43324e-05 (rank : 47) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8IWB7, Q9H9D5, Q9P2B3 | Gene names | WDFY1, KIAA1435, WDF1, ZFYVE17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 1 (WD40- and FYVE domain- containing protein 1) (Phosphoinositide-binding protein 1) (FENS-1) (Zinc finger FYVE domain-containing protein 17). | |||||
|
WDFY2_MOUSE
|
||||||
| NC score | 0.363861 (rank : 42) | θ value | 3.77169e-06 (rank : 43) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BUB4 | Gene names | Wdfy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 2 (WD40- and FYVE domain- containing protein 2). | |||||
|
WDFY2_HUMAN
|
||||||
| NC score | 0.361030 (rank : 43) | θ value | 3.77169e-06 (rank : 42) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96P53, Q96CS1 | Gene names | WDFY2, WDF2, ZFYVE22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 2 (WD40- and FYVE domain- containing protein 2) (Zinc finger FYVE domain-containing protein 22). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.298917 (rank : 44) | θ value | 3.89403e-11 (rank : 22) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
FYCO1_HUMAN
|
||||||
| NC score | 0.271554 (rank : 45) | θ value | 4.59992e-12 (rank : 15) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
ANFY1_MOUSE
|
||||||
| NC score | 0.252027 (rank : 46) | θ value | 2.36244e-08 (rank : 36) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q810B6, O54807, Q80TG6, Q80UH8 | Gene names | Ankfy1, Ankhzn, Kiaa1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
FYCO1_MOUSE
|
||||||
| NC score | 0.252002 (rank : 47) | θ value | 8.67504e-11 (rank : 25) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
ANFY1_HUMAN
|
||||||
| NC score | 0.246892 (rank : 48) | θ value | 4.45536e-07 (rank : 40) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9P2R3, Q9ULG5 | Gene names | ANKFY1, ANKHZN, KIAA1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.226768 (rank : 49) | θ value | 1.47974e-10 (rank : 27) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.209125 (rank : 50) | θ value | 4.30538e-10 (rank : 31) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
RFFL_MOUSE
|
||||||
| NC score | 0.187972 (rank : 51) | θ value | 0.0148317 (rank : 53) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6ZQM0, Q5SVC2, Q5SVC4, Q9D543, Q9D9B1 | Gene names | Rffl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rififylin (RING finger and FYVE-like domain-containing protein 1) (Fring). | |||||
|
RNF34_MOUSE
|
||||||
| NC score | 0.177785 (rank : 52) | θ value | 0.0431538 (rank : 56) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99KR6, Q3UV45 | Gene names | Rnf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 34 (RING finger protein RIFF) (Phafin-1). | |||||
|
RNF34_HUMAN
|
||||||
| NC score | 0.177297 (rank : 53) | θ value | 0.0330416 (rank : 55) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q969K3, Q8NG47, Q9H6W8 | Gene names | RNF34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 34 (RING finger protein RIFF) (FYVE-RING finger protein Momo) (Human RING finger homologous to inhibitor of apoptosis protein) (hRFI) (Caspases-8 and -10-associated RING finger protein 1) (CARP-1) (Caspase regulator CARP1). | |||||
|
RFFL_HUMAN
|
||||||
| NC score | 0.174657 (rank : 54) | θ value | 0.0563607 (rank : 57) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WZ73, Q8NHW0, Q8TBY7, Q96BE6 | Gene names | RFFL, RNF189, RNF34L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rififylin (RING finger and FYVE-like domain-containing protein 1) (FYVE-RING finger protein Sakura) (Fring) (Caspases-8 and -10- associated RING finger protein 2) (CARP-2) (Caspase regulator CARP2) (RING finger protein 189) (RING finger protein 34-like). | |||||
|
FARP2_HUMAN
|
||||||
| NC score | 0.140941 (rank : 55) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
FARP2_MOUSE
|
||||||
| NC score | 0.139686 (rank : 56) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
PREX1_HUMAN
|
||||||
| NC score | 0.138099 (rank : 57) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TCU6, Q5JS95, Q5JS96, Q69YL2, Q7Z2L9, Q9BQH0, Q9BX55, Q9H4Q6, Q9P2D2, Q9UGQ4 | Gene names | PREX1, KIAA1415 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (P-Rex1 protein). | |||||
|
FARP1_HUMAN
|
||||||
| NC score | 0.136383 (rank : 58) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
ECT2_MOUSE
|
||||||
| NC score | 0.132545 (rank : 59) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q07139 | Gene names | Ect2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
ECT2_HUMAN
|
||||||
| NC score | 0.129160 (rank : 60) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H8V3, Q9NSV8, Q9NVW9 | Gene names | ECT2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
RUFY3_MOUSE
|
||||||
| NC score | 0.117456 (rank : 61) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 579 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D394, Q3U348, Q3UF96, Q6PE64, Q8VD10 | Gene names | Rufy3, D5Bwg0860e, Ripx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RUFY3 (Rap2-interacting protein x) (RIPx). | |||||
|
RUFY3_HUMAN
|
||||||
| NC score | 0.116360 (rank : 62) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7L099, O94948, Q9UI00 | Gene names | RUFY3, KIAA0871 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RUFY3 (Rap2-interacting protein x) (RIPx). | |||||
|
PKHG1_HUMAN
|
||||||
| NC score | 0.115853 (rank : 63) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ULL1, Q5T1F2 | Gene names | PLEKHG1, KIAA1209 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family G member 1. | |||||
|
ARHG5_HUMAN
|
||||||
| NC score | 0.115256 (rank : 64) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q12774 | Gene names | ARHGEF5, TIM | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 5 (Guanine nucleotide regulatory protein TIM) (Oncogene TIM) (p60 TIM) (Transforming immortalized mammary oncogene). | |||||
|
ARHGA_MOUSE
|
||||||
| NC score | 0.109695 (rank : 65) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C033, Q5DU38, Q80VH8, Q8BW76, Q922S7 | Gene names | Arhgef10, Kiaa0294 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 10. | |||||
|
ARHGA_HUMAN
|
||||||
| NC score | 0.108949 (rank : 66) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15013, O14665, Q68D55, Q8IWD9, Q8IY77 | Gene names | ARHGEF10, KIAA0294 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 10. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.106399 (rank : 67) | θ value | 6.87365e-08 (rank : 37) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
HAPIP_HUMAN
|
||||||
| NC score | 0.105517 (rank : 68) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60229 | Gene names | HAPIP, DUO | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein-interacting protein (Protein Duo). | |||||
|
NGEF_HUMAN
|
||||||
| NC score | 0.102749 (rank : 69) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N5V2, Q53QQ4, Q53ST7, Q6GMQ5, Q9NQD6 | Gene names | NGEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephexin-1 (Eph-interacting exchange protein) (Neuronal guanine nucleotide exchange factor). | |||||
|
ARHGF_HUMAN
|
||||||
| NC score | 0.100205 (rank : 70) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O94989, Q8N449, Q9H8B4 | Gene names | ARHGEF15, KIAA0915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 15 (Vsm-RhoGEF). | |||||
|
ARHGG_HUMAN
|
||||||
| NC score | 0.099910 (rank : 71) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5VV41, Q86TF0, Q99434 | Gene names | ARHGEF16, NBR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
NGEF_MOUSE
|
||||||
| NC score | 0.099775 (rank : 72) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CHT1, Q8R204, Q923H2, Q9JHT9 | Gene names | Ngef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephexin-1 (Eph-interacting exchange protein) (Neuronal guanine nucleotide exchange factor). | |||||
|
ARHG2_MOUSE
|
||||||
| NC score | 0.093870 (rank : 73) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60875, O09115 | Gene names | Arhgef2, Lbcl1, Lfc | |||
|
Domain Architecture |

|
|||||
| Description | Rho/Rac guanine nucleotide exchange factor 2 (Lymphoid blast crisis- like 1) (LBC'S first cousin) (Oncogene LFC) (RHOBIN). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.093249 (rank : 74) | θ value | 0.00020696 (rank : 48) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ARHGG_MOUSE
|
||||||
| NC score | 0.092554 (rank : 75) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3U5C8, Q501M8, Q8VCE8 | Gene names | Arhgef16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
MCF2L_MOUSE
|
||||||
| NC score | 0.091079 (rank : 76) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q64096 | Gene names | Mcf2l, Dbs | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide exchange factor DBS (DBL's big sister) (MCF2- transforming sequence-like protein). | |||||
|
MCF2_HUMAN
|
||||||
| NC score | 0.090044 (rank : 77) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10911, P14919, Q5JYJ4, Q8IUF3, Q8IUF4, Q9UJB3 | Gene names | MCF2, DBL | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene DBL (Proto-oncogene MCF-2) [Contains: MCF2-transforming protein; DBL-transforming protein]. | |||||
|
PKHG4_HUMAN
|
||||||
| NC score | 0.088461 (rank : 78) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q58EX7, Q4G0J8, Q4H485, Q56A69, Q9H7K4, Q9UFW0 | Gene names | PLEKHG4, PRTPHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Puratrophin-1 (Pleckstrin homology domain-containing family G member 4) (Purkinje cell atrophy-associated protein 1). | |||||
|
LBC_HUMAN
|
||||||
| NC score | 0.086057 (rank : 79) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12802 | Gene names | LBC | |||
|
Domain Architecture |

|
|||||
| Description | LBC oncogene (P47) (Lymphoid blast crisis oncogene). | |||||
|
ARHG4_HUMAN
|
||||||
| NC score | 0.084947 (rank : 80) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NR80, Q9HDC6, Q9UPP0 | Gene names | ARHGEF4, KIAA1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
ARHG4_MOUSE
|
||||||
| NC score | 0.084254 (rank : 81) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TNR9, Q80TJ6 | Gene names | Arhgef4, Kiaa1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
GNRP_HUMAN
|
||||||
| NC score | 0.084251 (rank : 82) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
BCR_HUMAN
|
||||||
| NC score | 0.084035 (rank : 83) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
GNRP_MOUSE
|
||||||
| NC score | 0.083740 (rank : 84) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
ABR_HUMAN
|
||||||
| NC score | 0.082540 (rank : 85) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
ARHG1_HUMAN
|
||||||
| NC score | 0.082165 (rank : 86) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92888, O00513, Q8N4J4, Q96BF4, Q96F17, Q9BSB1 | Gene names | ARHGEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (p115-RhoGEF) (p115RhoGEF) (115 kDa guanine nucleotide exchange factor) (Sub1.5). | |||||
|
ARHG1_MOUSE
|
||||||
| NC score | 0.081413 (rank : 87) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
ARHG9_HUMAN
|
||||||
| NC score | 0.079060 (rank : 88) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43307, Q5JSL6 | Gene names | ARHGEF9, KIAA0424 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 9 (Rac/Cdc42 guanine nucleotide exchange factor 9) (Collybistin) (PEM-2 homolog). | |||||
|
CJ064_HUMAN
|
||||||
| NC score | 0.078945 (rank : 89) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZS81, Q8N4A3 | Gene names | C10orf64 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf64. | |||||
|
ARHG9_MOUSE
|
||||||
| NC score | 0.077734 (rank : 90) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3UTH8, Q3TQ60, Q80U06, Q8CAF9 | Gene names | Arhgef9, Kiaa0424 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 9 (Rac/Cdc42 guanine nucleotide exchange factor 9) (Collybistin). | |||||
|
ARHG2_HUMAN
|
||||||
| NC score | 0.068471 (rank : 91) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92974, O75142, Q15079 | Gene names | ARHGEF2, KIAA0651, LFP40 | |||
|
Domain Architecture |

|
|||||
| Description | Rho/Rac guanine nucleotide exchange factor 2 (GEF-H1 protein) (Proliferating cell nucleolar antigen p40). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.067815 (rank : 92) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.067752 (rank : 93) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
ITSN2_HUMAN
|
||||||
| NC score | 0.067523 (rank : 94) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
ITSN2_MOUSE
|
||||||
| NC score | 0.067498 (rank : 95) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
ARHGB_HUMAN
|
||||||
| NC score | 0.065621 (rank : 96) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
TIAM1_HUMAN
|
||||||
| NC score | 0.065588 (rank : 97) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13009 | Gene names | TIAM1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
ARHGC_HUMAN
|
||||||
| NC score | 0.064810 (rank : 98) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
ARHGC_MOUSE
|
||||||
| NC score | 0.064012 (rank : 99) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R4H2, Q80U18 | Gene names | Arhgef12, Kiaa0382, Larg | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
TIAM1_MOUSE
|
||||||
| NC score | 0.063631 (rank : 100) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60610 | Gene names | Tiam1, Tiam-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
ARHG7_HUMAN
|
||||||
| NC score | 0.063408 (rank : 101) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14155, Q6P9G3, Q6PII2, Q86W63, Q8N3M1 | Gene names | ARHGEF7, COOL1, KIAA0142, P85SPR, PAK3BP, PIXB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (COOL-1) (p85). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.063277 (rank : 102) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.060560 (rank : 103) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
VAV_HUMAN
|
||||||
| NC score | 0.056854 (rank : 104) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P15498, Q15860 | Gene names | VAV1, VAV | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene vav. | |||||
|
NBEAL_HUMAN
|
||||||
| NC score | 0.055177 (rank : 105) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZS30, Q6Y876, Q6ZQY5, Q96Q30 | Gene names | NBEAL1, ALS2CR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurobeachin-like 1 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 17 protein). | |||||
|
ARHG3_MOUSE
|
||||||
| NC score | 0.054645 (rank : 106) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91X46, Q8CDM0, Q91VY4, Q99K14, Q9DC31 | Gene names | Arhgef3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 3. | |||||
|
VAV_MOUSE
|
||||||
| NC score | 0.054320 (rank : 107) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P27870, Q8BTV7 | Gene names | Vav1, Vav | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene vav (p95vav). | |||||
|
SWP70_HUMAN
|
||||||
| NC score | 0.053265 (rank : 108) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
ALS2_MOUSE
|
||||||
| NC score | 0.053169 (rank : 109) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
|
ARHG8_HUMAN
|
||||||
| NC score | 0.053050 (rank : 110) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z628, Q12773, Q96D82, Q99903, Q9UEN6 | Gene names | NET1, ARHGEF8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroepithelial cell-transforming gene 1 protein (p65 Net1 proto- oncogene) (Rho guanine nucleotide exchange factor 8). | |||||
|
VAV2_MOUSE
|
||||||
| NC score | 0.053040 (rank : 111) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60992 | Gene names | Vav2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
ARHG3_HUMAN
|
||||||
| NC score | 0.052788 (rank : 112) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NR81, Q6NUN3, Q7Z4U2, Q7Z5T2, Q9H7T4 | Gene names | ARHGEF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 3 (Exchange factor found in platelets and leukemic and neuronal tissues) (XPLN). | |||||
|
ARHG7_MOUSE
|
||||||
| NC score | 0.052648 (rank : 113) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ES28, O08757, Q9ES27 | Gene names | Arhgef7, Pak3bp | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (p85SPR). | |||||
|
VAV3_HUMAN
|
||||||
| NC score | 0.051964 (rank : 114) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKW4, O95230, Q9Y5X8 | Gene names | VAV3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
VAV3_MOUSE
|
||||||
| NC score | 0.051291 (rank : 115) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9R0C8, Q7TS85 | Gene names | Vav3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
SWP70_MOUSE
|
||||||
| NC score | 0.050740 (rank : 116) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
WDR37_HUMAN
|
||||||
| NC score | 0.043672 (rank : 117) | θ value | 0.0563607 (rank : 58) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2I8, Q5SW03, Q9NTJ6 | Gene names | WDR37, KIAA0982 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 37. | |||||
|
WDR37_MOUSE
|
||||||
| NC score | 0.042458 (rank : 118) | θ value | 0.0563607 (rank : 59) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CBE3, Q80Y96, Q8CCL2 | Gene names | Wdr37, Kiaa0982 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 37. | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.037744 (rank : 119) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.037518 (rank : 120) | θ value | 1.06291 (rank : 70) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
AIM1_HUMAN
|
||||||
| NC score | 0.037478 (rank : 121) | θ value | 0.0961366 (rank : 61) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4K1, O00296, Q5VWJ2 | Gene names | AIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Absent in melanoma 1 protein. | |||||
|
IMA5_HUMAN
|
||||||
| NC score | 0.036438 (rank : 122) | θ value | 0.365318 (rank : 65) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15131 | Gene names | KPNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-6 subunit (Karyopherin alpha-5 subunit). | |||||
|
IMA1_HUMAN
|
||||||
| NC score | 0.035505 (rank : 123) | θ value | 0.813845 (rank : 68) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P52294, Q9BQ56 | Gene names | KPNA1, RCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (NPI-1). | |||||
|
NEB1_HUMAN
|
||||||
| NC score | 0.035261 (rank : 124) | θ value | 1.81305 (rank : 83) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.035018 (rank : 125) | θ value | 1.38821 (rank : 78) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
IMA1_MOUSE
|
||||||
| NC score | 0.034338 (rank : 126) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60960, Q3TF32 | Gene names | Kpna1, Rch2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (Importin alpha S1). | |||||
|
AFTIN_MOUSE
|
||||||
| NC score | 0.033877 (rank : 127) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80WT5, Q5SSE6, Q99KJ1 | Gene names | Aftph, Afth | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aftiphilin. | |||||
|
IMA7_MOUSE
|
||||||
| NC score | 0.033854 (rank : 128) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35345, Q9CVP9 | Gene names | Kpna6, Kpna5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6 subunit) (Importin alpha S2). | |||||
|
CEP57_MOUSE
|
||||||
| NC score | 0.033514 (rank : 129) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 580 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CEE0, Q6ZQJ3, Q7TN18, Q80X65, Q810F2, Q9D4J4, Q9D5S4, Q9D5W5 | Gene names | Cep57, Kiaa0092, Tsp57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin). | |||||
|
IMA7_HUMAN
|
||||||
| NC score | 0.033264 (rank : 130) | θ value | 1.81305 (rank : 82) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60684 | Gene names | KPNA6, IPOA7 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6). | |||||
|
GFRA2_MOUSE
|
||||||
| NC score | 0.032807 (rank : 131) | θ value | 0.163984 (rank : 62) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08842 | Gene names | Gfra2, Gdnfrb, Trnr2 | |||
|
Domain Architecture |

|
|||||
| Description | GDNF family receptor alpha-2 precursor (GFR-alpha-2) (Neurturin receptor alpha) (NTNR-alpha) (NRTNR-alpha) (TGF-beta-related neurotrophic factor receptor 2) (GDNF receptor beta) (GDNFR-beta). | |||||
|
BIN1_MOUSE
|
||||||
| NC score | 0.032009 (rank : 132) | θ value | 0.0736092 (rank : 60) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08539, Q62434 | Gene names | Bin1, Amphl, Sh3p9 | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (SH3-domain-containing protein 9). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.031062 (rank : 133) | θ value | 0.0252991 (rank : 54) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.030215 (rank : 134) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
SDPR_HUMAN
|
||||||
| NC score | 0.026276 (rank : 135) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95810 | Gene names | SDPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68). | |||||
|
GFRA2_HUMAN
|
||||||
| NC score | 0.023930 (rank : 136) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00451, O15316, O15328, Q7Z5C2 | Gene names | GFRA2, GDNFRB, RETL2, TRNR2 | |||
|
Domain Architecture |

|
|||||
| Description | GDNF family receptor alpha-2 precursor (GFR-alpha-2) (Neurturin receptor alpha) (NTNR-alpha) (NRTNR-alpha) (TGF-beta-related neurotrophic factor receptor 2) (GDNF receptor beta) (GDNFR-beta) (RET ligand 2). | |||||
|
PODXL_HUMAN
|
||||||
| NC score | 0.023732 (rank : 137) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00592 | Gene names | PODXL, PCLP, PCLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
SAS10_HUMAN
|
||||||
| NC score | 0.023112 (rank : 138) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQZ2, Q6FI82 | Gene names | CRLZ1, SAS10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Something about silencing protein 10 (Disrupter of silencing SAS10) (Charged amino acid-rich leucine zipper 1). | |||||
|
CND2_HUMAN
|
||||||
| NC score | 0.022077 (rank : 139) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15003, Q8TB87 | Gene names | NCAPH, BRRN, BRRN1, CAPH, KIAA0074 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 2 (Non-SMC condensin I complex subunit H) (Barren homolog protein 1) (Chromosome-associated protein H) (hCAP-H) (XCAP-H homolog). | |||||
|
MUC13_HUMAN
|
||||||
| NC score | 0.021589 (rank : 140) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3R2, Q6UWD9, Q9NXT5 | Gene names | MUC13, DRCC1, RECC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-13 precursor (Down-regulated in colon cancer 1). | |||||
|
DMRT1_MOUSE
|
||||||
| NC score | 0.021523 (rank : 141) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZ59, Q9QZ37, Q9QZA4 | Gene names | Dmrt1 | |||
|
Domain Architecture |

|
|||||
| Description | Doublesex- and mab-3-related transcription factor 1. | |||||
|
CND2_MOUSE
|
||||||
| NC score | 0.021074 (rank : 142) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C156, Q8BY01, Q8VDM9 | Gene names | Ncaph, Brrn, Brrn1, Caph | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 2 (Non-SMC condensin I complex subunit H) (Barren homolog protein 1) (Chromosome-associated protein H) (mCAP-H) (XCAP-H homolog). | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.020558 (rank : 143) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.019714 (rank : 144) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
K2027_HUMAN
|
||||||
| NC score | 0.019032 (rank : 145) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZVF9, Q8IVE4 | Gene names | KIAA2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
RFIP1_MOUSE
|
||||||
| NC score | 0.018946 (rank : 146) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D620, Q3UBC2, Q8BN24 | Gene names | Rab11fip1, Rcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.018639 (rank : 147) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
SCAR3_HUMAN
|
||||||
| NC score | 0.017501 (rank : 148) | θ value | 1.06291 (rank : 71) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6AZY7, Q9UM15, Q9UM16 | Gene names | SCARA3, CSR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class A member 3 (Cellular stress response gene protein). | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.017230 (rank : 149) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
CC14A_HUMAN
|
||||||
| NC score | 0.016739 (rank : 150) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UNH5, O43171, O60727, O60728, Q8IXX0 | Gene names | CDC14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
SAFB1_HUMAN
|
||||||
| NC score | 0.016166 (rank : 151) | θ value | 1.81305 (rank : 84) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15424, O60406, Q59HH8 | Gene names | SAFB, HAP, HET, SAFB1 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B (Scaffold attachment factor B1) (SAF-B) (HSP27 estrogen response element-TATA box-binding protein) (HSP27 ERE- TATA-binding protein). | |||||
|
EAF2_HUMAN
|
||||||
| NC score | 0.013667 (rank : 152) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96CJ1, Q9NZ82 | Gene names | EAF2, TRAITS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELL-associated factor 2 (Testosterone-regulated apoptosis inducer and tumor suppressor protein). | |||||
|
DLG7_HUMAN
|
||||||
| NC score | 0.012374 (rank : 153) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15398, Q8NG58 | Gene names | DLG7, KIAA0008 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 7 (Hepatoma up-regulated protein) (HURP). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.012086 (rank : 154) | θ value | 0.47712 (rank : 67) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
DCR1A_HUMAN
|
||||||
| NC score | 0.011607 (rank : 155) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PJP8, Q14701, Q6P5Y3, Q6PKL4 | Gene names | DCLRE1A, KIAA0086, SNM1, SNM1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA cross-link repair 1A protein (hSNM1) (hSNM1A). | |||||
|
ARID2_HUMAN
|
||||||
| NC score | 0.011502 (rank : 156) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
ZN295_HUMAN
|
||||||
| NC score | 0.011464 (rank : 157) | θ value | 0.21417 (rank : 63) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULJ3, Q6P4R0 | Gene names | ZNF295, KIAA1227, ZBTB21 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 295 (Zinc finger and BTB domain-containing protein 21). | |||||
|
DGKG_HUMAN
|
||||||
| NC score | 0.010160 (rank : 158) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49619 | Gene names | DGKG, DAGK3 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase gamma (EC 2.7.1.107) (Diglyceride kinase gamma) (DGK-gamma) (DAG kinase gamma). | |||||
|
SMYD5_MOUSE
|
||||||
| NC score | 0.009089 (rank : 159) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3TYX3, Q3TRB2, Q91YL6 | Gene names | Smyd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 5 (Protein NN8-4AG). | |||||
|
AFAD_HUMAN
|
||||||
| NC score | 0.008995 (rank : 160) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
CP4AC_MOUSE
|
||||||
| NC score | 0.008898 (rank : 161) | θ value | 0.365318 (rank : 64) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91WL5, Q3UNE4, Q6P931, Q8N7N3 | Gene names | Cyp4a12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 4A12 (EC 1.14.14.1) (CYPIVA12). | |||||
|
DGKG_MOUSE
|
||||||
| NC score | 0.008839 (rank : 162) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WG7 | Gene names | Dgkg, Dagk3 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase gamma (EC 2.7.1.107) (Diglyceride kinase gamma) (DGK-gamma) (DAG kinase gamma) (88 kDa diacylglycerol kinase). | |||||
|
RBP2_MOUSE
|
||||||
| NC score | 0.007807 (rank : 163) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ERU9, Q61992, Q8C9K9 | Gene names | Ranbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2 (RanBP2). | |||||
|
TRI37_MOUSE
|
||||||
| NC score | 0.007467 (rank : 164) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PCX9, Q8CHC5 | Gene names | Trim37, Kiaa0898 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 37. | |||||
|
WEE1B_HUMAN
|
||||||
| NC score | 0.007057 (rank : 165) | θ value | 0.365318 (rank : 66) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P0C1S8 | Gene names | WEE1B, WEE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wee1-like protein kinase 1B (EC 2.7.10.2) (Wee1B kinase). | |||||
|
FINC_MOUSE
|
||||||
| NC score | 0.007041 (rank : 166) | θ value | 1.81305 (rank : 80) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
ZO3_HUMAN
|
||||||
| NC score | 0.006627 (rank : 167) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95049 | Gene names | TJP3, ZO3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.006147 (rank : 168) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
SP2_HUMAN
|
||||||
| NC score | 0.005567 (rank : 169) | θ value | 0.813845 (rank : 69) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02086 | Gene names | SP2, KIAA0048 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp2. | |||||
|
MAGBI_HUMAN
|
||||||
| NC score | 0.005141 (rank : 170) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96M61 | Gene names | MAGEB18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen B18 (MAGE-B18 antigen). | |||||
|
WEE1B_MOUSE
|
||||||
| NC score | 0.004674 (rank : 171) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q66JT0, Q4U4S4, Q7TPV9 | Gene names | Wee1b, Wee2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wee1-like protein kinase 1B (EC 2.7.10.2) (Wee1B kinase) (mWee1B). | |||||
|
CNTP3_HUMAN
|
||||||
| NC score | 0.003569 (rank : 172) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZ76, Q9C0E9 | Gene names | CNTNAP3, CASPR3, KIAA1714 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 3 precursor (Cell recognition molecule Caspr3). | |||||
|
NR1H2_MOUSE
|
||||||
| NC score | 0.002197 (rank : 173) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60644 | Gene names | Nr1h2, Lxrb, Rip15, Unr, Unr2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterols receptor LXR-beta (Liver X receptor beta) (Nuclear orphan receptor LXR-beta) (Ubiquitously-expressed nuclear receptor) (Retinoid X receptor-interacting protein No.15). | |||||
|
PO2F1_HUMAN
|
||||||
| NC score | 0.001854 (rank : 174) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14859, Q5TBT7, Q6PK46, Q8NEU9, Q9BPV1 | Gene names | POU2F1, OCT1, OTF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 1 (Octamer-binding transcription factor 1) (Oct-1) (OTF-1) (NF-A1). | |||||
|
MAZ_HUMAN
|
||||||
| NC score | 0.001470 (rank : 175) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P56270, Q15703, Q99443 | Gene names | MAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myc-associated zinc finger protein (MAZI) (Purine-binding transcription factor) (Pur-1) (ZF87) (ZIF87). | |||||
|
MAZ_MOUSE
|
||||||
| NC score | 0.001432 (rank : 176) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P56671, Q9R1W0 | Gene names | Maz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myc-associated zinc finger protein (MAZI) (Purine-binding transcription factor) (Pur-1). | |||||
|
ZN408_HUMAN
|
||||||
| NC score | 0.001415 (rank : 177) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H9D4 | Gene names | ZNF408, PFM14, PRDM17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 408 (PR-domain zinc finger protein 17). | |||||
|
ZFY1_MOUSE
|
||||||
| NC score | 0.001067 (rank : 178) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10925 | Gene names | Zfy1, Zfy-1 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger Y-chromosomal protein 1. | |||||
|
ZN230_HUMAN
|
||||||
| NC score | 0.001034 (rank : 179) | θ value | 1.06291 (rank : 72) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UIE0, O15322, Q9P1U6 | Gene names | ZNF230, FDZF2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 230 (Zinc finger protein FDZF2). | |||||
|
PCD12_HUMAN
|
||||||
| NC score | 0.000995 (rank : 180) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NPG4, Q6UXB6, Q96KB8, Q9H7Y6, Q9H8E0 | Gene names | PCDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-12 precursor (Vascular cadherin-2) (Vascular endothelial cadherin-2) (VE-cadherin-2) (VE-cad-2). | |||||
|
VEZF1_HUMAN
|
||||||
| NC score | 0.000980 (rank : 181) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14119 | Gene names | VEZF1, DB1, ZNF161 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vascular endothelial zinc finger 1 (Zinc finger protein 161) (Putative transcription factor DB1). | |||||
|
ZN699_HUMAN
|
||||||
| NC score | 0.000932 (rank : 182) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q32M78, Q8N9A1 | Gene names | ZNF699 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 699 (Hangover homolog). | |||||
|
ZN317_HUMAN
|
||||||
| NC score | 0.000869 (rank : 183) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96PQ6, Q96PM0, Q96PM1, Q96PT2 | Gene names | ZNF317 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 317. | |||||
|
ZN555_HUMAN
|
||||||
| NC score | -0.000154 (rank : 184) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NEP9, Q8NA46, Q96MP1 | Gene names | ZNF555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 555. | |||||
|
CUBN_HUMAN
|
||||||
| NC score | -0.000156 (rank : 185) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60494, Q5VTA6, Q96RU9 | Gene names | CUBN, IFCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor) (Intrinsic factor-vitamin B12 receptor) (460 kDa receptor) (Intestinal intrinsic factor receptor). | |||||