Please be patient as the page loads
|
RCC1_HUMAN
|
||||||
| SwissProt Accessions | P18754 | Gene names | RCC1, CHC1 | |||
|
Domain Architecture |
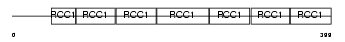
|
|||||
| Description | Regulator of chromosome condensation (Chromosome condensation protein 1) (Cell cycle regulatory protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RCC1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P18754 | Gene names | RCC1, CHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of chromosome condensation (Chromosome condensation protein 1) (Cell cycle regulatory protein). | |||||
|
RCC1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998432 (rank : 2) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8VE37, Q3UDB6 | Gene names | Rcc1, Chc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of chromosome condensation (Chromosome condensation protein 1). | |||||
|
HERC3_HUMAN
|
||||||
| θ value | 9.8567e-31 (rank : 3) | NC score | 0.609862 (rank : 17) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
RPGR_HUMAN
|
||||||
| θ value | 7.80994e-28 (rank : 4) | NC score | 0.805458 (rank : 7) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q92834, O00702, O00737, Q8N5T6, Q93039, Q9HD29, Q9UMR1 | Gene names | RPGR, RP3, XLRP3 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator. | |||||
|
HERC2_MOUSE
|
||||||
| θ value | 5.06226e-27 (rank : 5) | NC score | 0.610619 (rank : 16) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_HUMAN
|
||||||
| θ value | 8.63488e-27 (rank : 6) | NC score | 0.612900 (rank : 15) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
RPGR_MOUSE
|
||||||
| θ value | 8.08199e-25 (rank : 7) | NC score | 0.801233 (rank : 8) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
NEK9_MOUSE
|
||||||
| θ value | 5.79196e-23 (rank : 8) | NC score | 0.221539 (rank : 25) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 890 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8K1R7, Q8R3P1 | Gene names | Nek9, Nercc | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek9 (EC 2.7.11.1) (NimA-related protein kinase 9) (Never in mitosis A-related kinase 9). | |||||
|
NEK9_HUMAN
|
||||||
| θ value | 1.29031e-22 (rank : 9) | NC score | 0.218583 (rank : 26) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 900 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TD19, Q52LK6, Q8NCN0, Q8TCY4, Q9UPI4, Q9Y6S4, Q9Y6S5, Q9Y6S6 | Gene names | NEK9, KIAA1995, NEK8, NERCC | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek9 (EC 2.7.11.1) (NimA-related protein kinase 9) (Never in mitosis A-related kinase 9) (Nercc1 kinase) (NIMA-related kinase 8) (Nek8). | |||||
|
RCC2_HUMAN
|
||||||
| θ value | 2.87452e-22 (rank : 10) | NC score | 0.843845 (rank : 3) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9P258, Q8IVL9, Q9BSN6, Q9NPV8 | Gene names | RCC2, KIAA1470, TD60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2 (Telophase disk protein of 60 kDa) (RCC1-like protein TD- 60). | |||||
|
RCC2_MOUSE
|
||||||
| θ value | 3.75424e-22 (rank : 11) | NC score | 0.843376 (rank : 4) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BK67, Q6ZPQ0 | Gene names | Rcc2, Kiaa1470 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2. | |||||
|
SRGEF_HUMAN
|
||||||
| θ value | 1.33526e-19 (rank : 12) | NC score | 0.835366 (rank : 5) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UGK8, Q9UGK9 | Gene names | SERGEF, DELGEF, GNEFR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
SRGEF_MOUSE
|
||||||
| θ value | 2.5182e-18 (rank : 13) | NC score | 0.829841 (rank : 6) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80YD6, Q9QXB7 | Gene names | Sergef, Delgef, Gnefr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
WBS16_HUMAN
|
||||||
| θ value | 3.63628e-17 (rank : 14) | NC score | 0.800068 (rank : 9) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96I51, Q548B1, Q9H0G7 | Gene names | WBSCR16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Williams-Beuren syndrome chromosome region 16 protein (RCC1-like G exchanging factor-like protein). | |||||
|
RCBT1_HUMAN
|
||||||
| θ value | 4.74913e-17 (rank : 15) | NC score | 0.654861 (rank : 13) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NDN9, Q969U9 | Gene names | RCBTB1, CLLD7, E4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 1 (Regulator of chromosome condensation and BTB domain-containing protein 1) (Chronic lymphocytic leukemia deletion region gene 7 protein) (CLL deletion region gene 7 protein). | |||||
|
RCBT1_MOUSE
|
||||||
| θ value | 1.058e-16 (rank : 16) | NC score | 0.652198 (rank : 14) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6NXM2, Q8BTZ6, Q8BZV0 | Gene names | Rcbtb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 1 (Regulator of chromosome condensation and BTB domain-containing protein 1). | |||||
|
HERC5_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 17) | NC score | 0.576616 (rank : 18) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UII4 | Gene names | HERC5, CEB1, CEBP1 | |||
|
Domain Architecture |

|
|||||
| Description | HECT domain and RCC1-like domain-containing protein 5 (Cyclin-E- binding protein 1). | |||||
|
RCBT2_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 18) | NC score | 0.674739 (rank : 12) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99LJ7, Q3TUA3, Q8BMG2 | Gene names | Rcbtb2, Chc1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 2 (Regulator of chromosome condensation and BTB domain-containing protein 2) (Chromosome condensation 1-like). | |||||
|
NEK8_MOUSE
|
||||||
| θ value | 4.44505e-15 (rank : 19) | NC score | 0.200137 (rank : 27) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91ZR4, Q3U498, Q9D685 | Gene names | Nek8, Jck | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek8 (EC 2.7.11.1) (Never in mitosis A-related kinase 8) (NimA-related protein kinase 8). | |||||
|
RCBT2_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 20) | NC score | 0.680142 (rank : 11) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O95199 | Gene names | RCBTB2, CHC1L, RLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 2 (Regulator of chromosome condensation and BTB domain-containing protein 2) (Chromosome condensation 1-like) (CHC1-L) (RCC1-like G exchanging factor). | |||||
|
NEK8_HUMAN
|
||||||
| θ value | 4.91457e-14 (rank : 21) | NC score | 0.198821 (rank : 28) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q86SG6, Q2M1S6, Q8NDH1 | Gene names | NEK8, JCK, NEK12A | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek8 (EC 2.7.11.1) (Never in mitosis A-related kinase 8) (NimA-related protein kinase 8) (NIMA-related kinase 12a). | |||||
|
WBS16_MOUSE
|
||||||
| θ value | 4.91457e-14 (rank : 22) | NC score | 0.784213 (rank : 10) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CYF5 | Gene names | Wbscr16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Williams-Beuren syndrome chromosome region 16 protein homolog. | |||||
|
ALS2_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 23) | NC score | 0.520910 (rank : 19) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
|
ALS2_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 24) | NC score | 0.518430 (rank : 20) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
MYCB2_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 25) | NC score | 0.423350 (rank : 21) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
MYCB2_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 26) | NC score | 0.419300 (rank : 22) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
FBX24_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 27) | NC score | 0.327279 (rank : 23) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75426, Q9H0G1 | Gene names | FBXO24, FBX24 | |||
|
Domain Architecture |
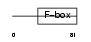
|
|||||
| Description | F-box only protein 24. | |||||
|
FBX24_MOUSE
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.287611 (rank : 24) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D417 | Gene names | Fbxo24, Fbx24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 24. | |||||
|
PCDA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.032914 (rank : 77) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y5I3, O75288, Q9NRT7 | Gene names | PCDHA1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 1 precursor (PCDH-alpha1). | |||||
|
PCDA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.032017 (rank : 83) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y5H9, O75287, Q9BTV3 | Gene names | PCDHA2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 2 precursor (PCDH-alpha2). | |||||
|
PCDA3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.032939 (rank : 76) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y5H8, O75286 | Gene names | PCDHA3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 3 precursor (PCDH-alpha3). | |||||
|
PCDA4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.033079 (rank : 75) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UN74, O75285 | Gene names | PCDHA4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 4 precursor (PCDH-alpha4). | |||||
|
PCDA4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.032047 (rank : 82) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88689, Q3UEX3, Q6PAM9, Q8K487, Q8K489 | Gene names | Pcdha4, Cnr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin alpha 4 precursor (PCDH-alpha4). | |||||
|
PCDA5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.031992 (rank : 84) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y5H7, O75284, Q8N4R3 | Gene names | PCDHA5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 5 precursor (PCDH-alpha5). | |||||
|
PCDA6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.033844 (rank : 71) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UN73, O75283, Q9NRT8 | Gene names | PCDHA6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 6 precursor (PCDH-alpha6). | |||||
|
PCDA7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.032869 (rank : 78) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UN72, O75282 | Gene names | PCDHA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 7 precursor (PCDH-alpha7). | |||||
|
PCDA8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.033422 (rank : 73) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y5H6, O75281 | Gene names | PCDHA8 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 8 precursor (PCDH-alpha8). | |||||
|
PCDA9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.033426 (rank : 72) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y5H5, O15053 | Gene names | PCDHA9, KIAA0345 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 9 precursor (PCDH-alpha9). | |||||
|
PCDAA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.033134 (rank : 74) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y5I2, O75280, Q9NRU2 | Gene names | PCDHA10 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 10 precursor (PCDH-alpha10). | |||||
|
PCDAB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.032829 (rank : 79) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y5I1, O75279 | Gene names | PCDHA11 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 11 precursor (PCDH-alpha11). | |||||
|
PCDAC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.032460 (rank : 80) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UN75, O75278 | Gene names | PCDHA12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 12 precursor (PCDH-alpha12). | |||||
|
PCDAD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.032277 (rank : 81) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y5I0, O75277 | Gene names | PCDHA13 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 13 precursor (PCDH-alpha13). | |||||
|
PCDC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.029125 (rank : 85) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H158, Q9Y5F5, Q9Y5I5 | Gene names | PCDHAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha C1 precursor (PCDH-alpha-C1). | |||||
|
PCDC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.028023 (rank : 86) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y5I4, Q9Y5F4 | Gene names | PCDHAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha C2 precursor (PCDH-alpha-C2). | |||||
|
GBRA3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.003591 (rank : 89) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P26049 | Gene names | Gabra3, Gabra-3 | |||
|
Domain Architecture |
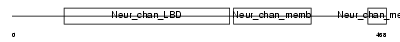
|
|||||
| Description | Gamma-aminobutyric-acid receptor subunit alpha-3 precursor (GABA(A) receptor subunit alpha-3). | |||||
|
MORG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.008213 (rank : 87) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DAJ4, Q505C2, Q8VEB7, Q99JX9, Q9D235 | Gene names | Morg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase organizer 1 (MAPK organizer 1). | |||||
|
CTR2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.006243 (rank : 88) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P18581 | Gene names | Slc7a2, Atrc2, Tea | |||
|
Domain Architecture |

|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2) (TEA protein) (T-cell early activation protein) (20.5). | |||||
|
ABTB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.052070 (rank : 63) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N961, Q6MZW4, Q8NB44 | Gene names | ABTB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
ABTB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.052060 (rank : 64) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TQI7 | Gene names | Abtb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
BTBD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.050063 (rank : 70) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BX70, O60418, O75248, Q7Z5W0, Q96SX8, Q9NPS1, Q9NX81 | Gene names | BTBD2 | |||
|
Domain Architecture |
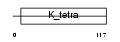
|
|||||
| Description | BTB/POZ domain-containing protein 2. | |||||
|
BTBD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.051547 (rank : 67) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y2F9 | Gene names | BTBD3, KIAA0952 | |||
|
Domain Architecture |
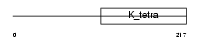
|
|||||
| Description | BTB/POZ domain-containing protein 3. | |||||
|
BTBD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.052017 (rank : 65) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P58545 | Gene names | Btbd3 | |||
|
Domain Architecture |
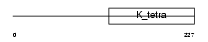
|
|||||
| Description | BTB/POZ domain-containing protein 3. | |||||
|
BTBD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.061057 (rank : 59) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5XKL5, Q6V9S5 | Gene names | BTBD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 8. | |||||
|
BTBDC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.056782 (rank : 62) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
CUL7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.064102 (rank : 58) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14999, Q5T654 | Gene names | CUL7, KIAA0076 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-7 (CUL-7). | |||||
|
EDD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.080348 (rank : 47) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
EDD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.079600 (rank : 50) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
HECD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.068098 (rank : 57) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
HECD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.109711 (rank : 33) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5U5R9, Q5VZ97, Q5VZ99, Q8TCP5 | Gene names | HECTD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.108251 (rank : 34) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CDU6, Q8CBQ9 | Gene names | Hectd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.125704 (rank : 29) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T447, Q5T448, Q9H783 | Gene names | HECTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HECD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.125641 (rank : 30) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3U487, Q3TN76, Q641P3, Q8BQ74, Q8R1L6 | Gene names | Hectd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.087768 (rank : 39) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.088867 (rank : 38) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
ITCH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.084280 (rank : 44) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
ITCH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.084939 (rank : 43) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
K0317_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.074010 (rank : 55) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15033, Q7LDY1, Q8IYY9 | Gene names | KIAA0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
K0317_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.073956 (rank : 56) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHG5, Q6P9Q1, Q80YC4, Q8C5W5 | Gene names | Kiaa0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
LZTR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.056941 (rank : 60) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N653, Q14776 | Gene names | LZTR1, TCFL2 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-zipper-like transcriptional regulator 1 (LZTR-1). | |||||
|
LZTR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.056853 (rank : 61) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CQ33 | Gene names | Lztr1, Tcfl2 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-zipper-like transcriptional regulator 1 (LZTR-1). | |||||
|
NED4L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.078182 (rank : 53) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NED4L_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.078301 (rank : 52) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NEDD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.077340 (rank : 54) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
NEDD4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.078974 (rank : 51) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
PARC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.050808 (rank : 69) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80TT8, Q8BKL9, Q8CGC0, Q8R363 | Gene names | Parc, Kiaa0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein. | |||||
|
RHBT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.051590 (rank : 66) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94955, Q8IW06 | Gene names | RHOBTB3, KIAA0878 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-related BTB domain-containing protein 3. | |||||
|
RHBT3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.051352 (rank : 68) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CTN4, Q80X55, Q9CVT0 | Gene names | Rhobtb3, Kiaa0878 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-related BTB domain-containing protein 3. | |||||
|
SMUF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.086157 (rank : 41) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
SMUF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.085763 (rank : 42) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
SMUF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.086299 (rank : 40) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.093458 (rank : 35) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
UBE3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.111604 (rank : 31) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
UBE3A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.110821 (rank : 32) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08759, P97482 | Gene names | Ube3a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (Oncogenic protein- associated protein E6-AP). | |||||
|
UBE3C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.093434 (rank : 36) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UBE3C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.093180 (rank : 37) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
WWP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.080111 (rank : 49) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
WWP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.081059 (rank : 46) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
WWP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.080307 (rank : 48) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
WWP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.081952 (rank : 45) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
RCC1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P18754 | Gene names | RCC1, CHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of chromosome condensation (Chromosome condensation protein 1) (Cell cycle regulatory protein). | |||||
|
RCC1_MOUSE
|
||||||
| NC score | 0.998432 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8VE37, Q3UDB6 | Gene names | Rcc1, Chc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of chromosome condensation (Chromosome condensation protein 1). | |||||
|
RCC2_HUMAN
|
||||||
| NC score | 0.843845 (rank : 3) | θ value | 2.87452e-22 (rank : 10) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9P258, Q8IVL9, Q9BSN6, Q9NPV8 | Gene names | RCC2, KIAA1470, TD60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2 (Telophase disk protein of 60 kDa) (RCC1-like protein TD- 60). | |||||
|
RCC2_MOUSE
|
||||||
| NC score | 0.843376 (rank : 4) | θ value | 3.75424e-22 (rank : 11) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BK67, Q6ZPQ0 | Gene names | Rcc2, Kiaa1470 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2. | |||||
|
SRGEF_HUMAN
|
||||||
| NC score | 0.835366 (rank : 5) | θ value | 1.33526e-19 (rank : 12) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UGK8, Q9UGK9 | Gene names | SERGEF, DELGEF, GNEFR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
SRGEF_MOUSE
|
||||||
| NC score | 0.829841 (rank : 6) | θ value | 2.5182e-18 (rank : 13) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80YD6, Q9QXB7 | Gene names | Sergef, Delgef, Gnefr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
RPGR_HUMAN
|
||||||
| NC score | 0.805458 (rank : 7) | θ value | 7.80994e-28 (rank : 4) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q92834, O00702, O00737, Q8N5T6, Q93039, Q9HD29, Q9UMR1 | Gene names | RPGR, RP3, XLRP3 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator. | |||||
|
RPGR_MOUSE
|
||||||
| NC score | 0.801233 (rank : 8) | θ value | 8.08199e-25 (rank : 7) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
WBS16_HUMAN
|
||||||
| NC score | 0.800068 (rank : 9) | θ value | 3.63628e-17 (rank : 14) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96I51, Q548B1, Q9H0G7 | Gene names | WBSCR16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Williams-Beuren syndrome chromosome region 16 protein (RCC1-like G exchanging factor-like protein). | |||||
|
WBS16_MOUSE
|
||||||
| NC score | 0.784213 (rank : 10) | θ value | 4.91457e-14 (rank : 22) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CYF5 | Gene names | Wbscr16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Williams-Beuren syndrome chromosome region 16 protein homolog. | |||||
|
RCBT2_HUMAN
|
||||||
| NC score | 0.680142 (rank : 11) | θ value | 2.20605e-14 (rank : 20) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O95199 | Gene names | RCBTB2, CHC1L, RLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 2 (Regulator of chromosome condensation and BTB domain-containing protein 2) (Chromosome condensation 1-like) (CHC1-L) (RCC1-like G exchanging factor). | |||||
|
RCBT2_MOUSE
|
||||||
| NC score | 0.674739 (rank : 12) | θ value | 3.40345e-15 (rank : 18) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99LJ7, Q3TUA3, Q8BMG2 | Gene names | Rcbtb2, Chc1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 2 (Regulator of chromosome condensation and BTB domain-containing protein 2) (Chromosome condensation 1-like). | |||||
|
RCBT1_HUMAN
|
||||||
| NC score | 0.654861 (rank : 13) | θ value | 4.74913e-17 (rank : 15) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NDN9, Q969U9 | Gene names | RCBTB1, CLLD7, E4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 1 (Regulator of chromosome condensation and BTB domain-containing protein 1) (Chronic lymphocytic leukemia deletion region gene 7 protein) (CLL deletion region gene 7 protein). | |||||
|
RCBT1_MOUSE
|
||||||
| NC score | 0.652198 (rank : 14) | θ value | 1.058e-16 (rank : 16) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6NXM2, Q8BTZ6, Q8BZV0 | Gene names | Rcbtb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 1 (Regulator of chromosome condensation and BTB domain-containing protein 1). | |||||
|
HERC2_HUMAN
|
||||||
| NC score | 0.612900 (rank : 15) | θ value | 8.63488e-27 (rank : 6) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_MOUSE
|
||||||
| NC score | 0.610619 (rank : 16) | θ value | 5.06226e-27 (rank : 5) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC3_HUMAN
|
||||||
| NC score | 0.609862 (rank : 17) | θ value | 9.8567e-31 (rank : 3) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
HERC5_HUMAN
|
||||||
| NC score | 0.576616 (rank : 18) | θ value | 3.40345e-15 (rank : 17) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UII4 | Gene names | HERC5, CEB1, CEBP1 | |||
|
Domain Architecture |

|
|||||
| Description | HECT domain and RCC1-like domain-containing protein 5 (Cyclin-E- binding protein 1). | |||||
|
ALS2_MOUSE
|
||||||
| NC score | 0.520910 (rank : 19) | θ value | 1.17247e-07 (rank : 23) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
|
ALS2_HUMAN
|
||||||
| NC score | 0.518430 (rank : 20) | θ value | 7.59969e-07 (rank : 24) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
MYCB2_HUMAN
|
||||||
| NC score | 0.423350 (rank : 21) | θ value | 7.59969e-07 (rank : 25) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
MYCB2_MOUSE
|
||||||
| NC score | 0.419300 (rank : 22) | θ value | 8.40245e-06 (rank : 26) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
FBX24_HUMAN
|
||||||
| NC score | 0.327279 (rank : 23) | θ value | 0.0113563 (rank : 27) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75426, Q9H0G1 | Gene names | FBXO24, FBX24 | |||
|
Domain Architecture |

|
|||||
| Description | F-box only protein 24. | |||||
|
FBX24_MOUSE
|
||||||
| NC score | 0.287611 (rank : 24) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D417 | Gene names | Fbxo24, Fbx24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 24. | |||||
|
NEK9_MOUSE
|
||||||
| NC score | 0.221539 (rank : 25) | θ value | 5.79196e-23 (rank : 8) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 890 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8K1R7, Q8R3P1 | Gene names | Nek9, Nercc | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek9 (EC 2.7.11.1) (NimA-related protein kinase 9) (Never in mitosis A-related kinase 9). | |||||
|
NEK9_HUMAN
|
||||||
| NC score | 0.218583 (rank : 26) | θ value | 1.29031e-22 (rank : 9) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 900 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TD19, Q52LK6, Q8NCN0, Q8TCY4, Q9UPI4, Q9Y6S4, Q9Y6S5, Q9Y6S6 | Gene names | NEK9, KIAA1995, NEK8, NERCC | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek9 (EC 2.7.11.1) (NimA-related protein kinase 9) (Never in mitosis A-related kinase 9) (Nercc1 kinase) (NIMA-related kinase 8) (Nek8). | |||||
|
NEK8_MOUSE
|
||||||
| NC score | 0.200137 (rank : 27) | θ value | 4.44505e-15 (rank : 19) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91ZR4, Q3U498, Q9D685 | Gene names | Nek8, Jck | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek8 (EC 2.7.11.1) (Never in mitosis A-related kinase 8) (NimA-related protein kinase 8). | |||||
|
NEK8_HUMAN
|
||||||
| NC score | 0.198821 (rank : 28) | θ value | 4.91457e-14 (rank : 21) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q86SG6, Q2M1S6, Q8NDH1 | Gene names | NEK8, JCK, NEK12A | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek8 (EC 2.7.11.1) (Never in mitosis A-related kinase 8) (NimA-related protein kinase 8) (NIMA-related kinase 12a). | |||||
|
HECD3_HUMAN
|
||||||
| NC score | 0.125704 (rank : 29) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T447, Q5T448, Q9H783 | Gene names | HECTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HECD3_MOUSE
|
||||||
| NC score | 0.125641 (rank : 30) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3U487, Q3TN76, Q641P3, Q8BQ74, Q8R1L6 | Gene names | Hectd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
UBE3A_HUMAN
|
||||||
| NC score | 0.111604 (rank : 31) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
UBE3A_MOUSE
|
||||||
| NC score | 0.110821 (rank : 32) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08759, P97482 | Gene names | Ube3a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (Oncogenic protein- associated protein E6-AP). | |||||
|
HECD2_HUMAN
|
||||||
| NC score | 0.109711 (rank : 33) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5U5R9, Q5VZ97, Q5VZ99, Q8TCP5 | Gene names | HECTD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD2_MOUSE
|
||||||
| NC score | 0.108251 (rank : 34) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CDU6, Q8CBQ9 | Gene names | Hectd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.093458 (rank : 35) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
UBE3C_HUMAN
|
||||||
| NC score | 0.093434 (rank : 36) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UBE3C_MOUSE
|
||||||
| NC score | 0.093180 (rank : 37) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.088867 (rank : 38) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.087768 (rank : 39) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
SMUF2_HUMAN
|
||||||
| NC score | 0.086299 (rank : 40) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
SMUF1_HUMAN
|
||||||
| NC score | 0.086157 (rank : 41) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
SMUF1_MOUSE
|
||||||
| NC score | 0.085763 (rank : 42) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
ITCH_MOUSE
|
||||||
| NC score | 0.084939 (rank : 43) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
ITCH_HUMAN
|
||||||
| NC score | 0.084280 (rank : 44) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
WWP2_MOUSE
|
||||||
| NC score | 0.081952 (rank : 45) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
WWP1_MOUSE
|
||||||
| NC score | 0.081059 (rank : 46) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
EDD1_HUMAN
|
||||||
| NC score | 0.080348 (rank : 47) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
WWP2_HUMAN
|
||||||
| NC score | 0.080307 (rank : 48) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
WWP1_HUMAN
|
||||||
| NC score | 0.080111 (rank : 49) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
EDD1_MOUSE
|
||||||
| NC score | 0.079600 (rank : 50) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
NEDD4_MOUSE
|
||||||
| NC score | 0.078974 (rank : 51) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
NED4L_MOUSE
|
||||||
| NC score | 0.078301 (rank : 52) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NED4L_HUMAN
|
||||||
| NC score | 0.078182 (rank : 53) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NEDD4_HUMAN
|
||||||
| NC score | 0.077340 (rank : 54) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
K0317_HUMAN
|
||||||
| NC score | 0.074010 (rank : 55) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15033, Q7LDY1, Q8IYY9 | Gene names | KIAA0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
K0317_MOUSE
|
||||||
| NC score | 0.073956 (rank : 56) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHG5, Q6P9Q1, Q80YC4, Q8C5W5 | Gene names | Kiaa0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
HECD1_HUMAN
|
||||||
| NC score | 0.068098 (rank : 57) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
CUL7_HUMAN
|
||||||
| NC score | 0.064102 (rank : 58) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14999, Q5T654 | Gene names | CUL7, KIAA0076 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-7 (CUL-7). | |||||
|
BTBD8_HUMAN
|
||||||
| NC score | 0.061057 (rank : 59) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5XKL5, Q6V9S5 | Gene names | BTBD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 8. | |||||
|
LZTR1_HUMAN
|
||||||
| NC score | 0.056941 (rank : 60) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N653, Q14776 | Gene names | LZTR1, TCFL2 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-zipper-like transcriptional regulator 1 (LZTR-1). | |||||
|
LZTR1_MOUSE
|
||||||
| NC score | 0.056853 (rank : 61) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CQ33 | Gene names | Lztr1, Tcfl2 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-zipper-like transcriptional regulator 1 (LZTR-1). | |||||
|
BTBDC_HUMAN
|
||||||
| NC score | 0.056782 (rank : 62) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
ABTB2_HUMAN
|
||||||
| NC score | 0.052070 (rank : 63) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N961, Q6MZW4, Q8NB44 | Gene names | ABTB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
ABTB2_MOUSE
|
||||||
| NC score | 0.052060 (rank : 64) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TQI7 | Gene names | Abtb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
BTBD3_MOUSE
|
||||||
| NC score | 0.052017 (rank : 65) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P58545 | Gene names | Btbd3 | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein 3. | |||||
|
RHBT3_HUMAN
|
||||||
| NC score | 0.051590 (rank : 66) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94955, Q8IW06 | Gene names | RHOBTB3, KIAA0878 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-related BTB domain-containing protein 3. | |||||
|
BTBD3_HUMAN
|
||||||
| NC score | 0.051547 (rank : 67) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y2F9 | Gene names | BTBD3, KIAA0952 | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein 3. | |||||
|
RHBT3_MOUSE
|
||||||
| NC score | 0.051352 (rank : 68) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CTN4, Q80X55, Q9CVT0 | Gene names | Rhobtb3, Kiaa0878 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-related BTB domain-containing protein 3. | |||||
|
PARC_MOUSE
|
||||||
| NC score | 0.050808 (rank : 69) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80TT8, Q8BKL9, Q8CGC0, Q8R363 | Gene names | Parc, Kiaa0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein. | |||||
|
BTBD2_HUMAN
|
||||||
| NC score | 0.050063 (rank : 70) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BX70, O60418, O75248, Q7Z5W0, Q96SX8, Q9NPS1, Q9NX81 | Gene names | BTBD2 | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein 2. | |||||
|
PCDA6_HUMAN
|
||||||
| NC score | 0.033844 (rank : 71) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UN73, O75283, Q9NRT8 | Gene names | PCDHA6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 6 precursor (PCDH-alpha6). | |||||
|
PCDA9_HUMAN
|
||||||
| NC score | 0.033426 (rank : 72) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y5H5, O15053 | Gene names | PCDHA9, KIAA0345 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 9 precursor (PCDH-alpha9). | |||||
|
PCDA8_HUMAN
|
||||||
| NC score | 0.033422 (rank : 73) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y5H6, O75281 | Gene names | PCDHA8 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 8 precursor (PCDH-alpha8). | |||||
|
PCDAA_HUMAN
|
||||||
| NC score | 0.033134 (rank : 74) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y5I2, O75280, Q9NRU2 | Gene names | PCDHA10 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 10 precursor (PCDH-alpha10). | |||||
|
PCDA4_HUMAN
|
||||||
| NC score | 0.033079 (rank : 75) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UN74, O75285 | Gene names | PCDHA4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 4 precursor (PCDH-alpha4). | |||||
|
PCDA3_HUMAN
|
||||||
| NC score | 0.032939 (rank : 76) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y5H8, O75286 | Gene names | PCDHA3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 3 precursor (PCDH-alpha3). | |||||
|
PCDA1_HUMAN
|
||||||
| NC score | 0.032914 (rank : 77) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y5I3, O75288, Q9NRT7 | Gene names | PCDHA1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 1 precursor (PCDH-alpha1). | |||||
|
PCDA7_HUMAN
|
||||||
| NC score | 0.032869 (rank : 78) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UN72, O75282 | Gene names | PCDHA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 7 precursor (PCDH-alpha7). | |||||
|
PCDAB_HUMAN
|
||||||
| NC score | 0.032829 (rank : 79) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y5I1, O75279 | Gene names | PCDHA11 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 11 precursor (PCDH-alpha11). | |||||
|
PCDAC_HUMAN
|
||||||
| NC score | 0.032460 (rank : 80) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UN75, O75278 | Gene names | PCDHA12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 12 precursor (PCDH-alpha12). | |||||
|
PCDAD_HUMAN
|
||||||
| NC score | 0.032277 (rank : 81) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y5I0, O75277 | Gene names | PCDHA13 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 13 precursor (PCDH-alpha13). | |||||
|
PCDA4_MOUSE
|
||||||
| NC score | 0.032047 (rank : 82) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88689, Q3UEX3, Q6PAM9, Q8K487, Q8K489 | Gene names | Pcdha4, Cnr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin alpha 4 precursor (PCDH-alpha4). | |||||
|
PCDA2_HUMAN
|
||||||
| NC score | 0.032017 (rank : 83) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y5H9, O75287, Q9BTV3 | Gene names | PCDHA2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 2 precursor (PCDH-alpha2). | |||||
|
PCDA5_HUMAN
|
||||||
| NC score | 0.031992 (rank : 84) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y5H7, O75284, Q8N4R3 | Gene names | PCDHA5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 5 precursor (PCDH-alpha5). | |||||
|
PCDC1_HUMAN
|
||||||
| NC score | 0.029125 (rank : 85) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H158, Q9Y5F5, Q9Y5I5 | Gene names | PCDHAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha C1 precursor (PCDH-alpha-C1). | |||||
|
PCDC2_HUMAN
|
||||||
| NC score | 0.028023 (rank : 86) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y5I4, Q9Y5F4 | Gene names | PCDHAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha C2 precursor (PCDH-alpha-C2). | |||||
|
MORG1_MOUSE
|
||||||
| NC score | 0.008213 (rank : 87) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DAJ4, Q505C2, Q8VEB7, Q99JX9, Q9D235 | Gene names | Morg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase organizer 1 (MAPK organizer 1). | |||||
|
CTR2_MOUSE
|
||||||
| NC score | 0.006243 (rank : 88) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P18581 | Gene names | Slc7a2, Atrc2, Tea | |||
|
Domain Architecture |

|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2) (TEA protein) (T-cell early activation protein) (20.5). | |||||
|
GBRA3_MOUSE
|
||||||
| NC score | 0.003591 (rank : 89) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P26049 | Gene names | Gabra3, Gabra-3 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric-acid receptor subunit alpha-3 precursor (GABA(A) receptor subunit alpha-3). | |||||