Please be patient as the page loads
|
JPH1_MOUSE
|
||||||
| SwissProt Accessions | Q9ET80, Q9EQZ3 | Gene names | Jph1, Jp1 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
JPH1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.987102 (rank : 2) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9HDC5 | Gene names | JPH1, JP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
JPH1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 120 | |
| SwissProt Accessions | Q9ET80, Q9EQZ3 | Gene names | Jph1, Jp1 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 1.24043e-158 (rank : 3) | NC score | 0.877916 (rank : 8) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
JPH2_MOUSE
|
||||||
| θ value | 6.37065e-155 (rank : 4) | NC score | 0.936563 (rank : 7) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
JPH3_HUMAN
|
||||||
| θ value | 1.20145e-153 (rank : 5) | NC score | 0.968144 (rank : 3) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WXH2, Q8N471, Q9HDC3, Q9HDC4 | Gene names | JPH3, JP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
JPH3_MOUSE
|
||||||
| θ value | 4.56552e-153 (rank : 6) | NC score | 0.967876 (rank : 4) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ET77, Q9EQZ2 | Gene names | Jph3, Jp3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
JPH4_MOUSE
|
||||||
| θ value | 4.95767e-99 (rank : 7) | NC score | 0.944674 (rank : 6) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q80WT0, Q8BMI1 | Gene names | Jph4, Jphl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
JPH4_HUMAN
|
||||||
| θ value | 2.46047e-98 (rank : 8) | NC score | 0.946048 (rank : 5) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96JJ6, Q8ND44, Q96DQ0 | Gene names | JPH4, JPHL1, KIAA1831 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
MORN3_MOUSE
|
||||||
| θ value | 5.43371e-13 (rank : 9) | NC score | 0.673943 (rank : 9) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8C5T4, Q8VE21, Q9D5H6 | Gene names | Morn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 3. | |||||
|
MORN3_HUMAN
|
||||||
| θ value | 9.26847e-13 (rank : 10) | NC score | 0.665912 (rank : 10) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PF18, Q86YQ9 | Gene names | MORN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 3. | |||||
|
TSGA2_MOUSE
|
||||||
| θ value | 6.00763e-12 (rank : 11) | NC score | 0.645466 (rank : 12) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VIG3, Q9DAL5 | Gene names | Tsga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific gene A2 protein (Male meiotic metaphase chromosome- associated acidic protein) (Meichroacidin). | |||||
|
TSGA2_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 12) | NC score | 0.652526 (rank : 11) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WYR4 | Gene names | TSGA2, TSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific gene A2 protein (Male meiotic metaphase chromosome- associated acidic protein) (Meichroacidin). | |||||
|
MORN1_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 13) | NC score | 0.622742 (rank : 13) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5T089, Q8WW30, Q9H852 | Gene names | MORN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 1. | |||||
|
ALS2_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 14) | NC score | 0.413604 (rank : 16) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
ALS2_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 15) | NC score | 0.413582 (rank : 17) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
|
MORN2_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 16) | NC score | 0.517877 (rank : 14) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q502X0, Q6UL00 | Gene names | MORN2, MOPT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 2 (MORN motif protein in testis). | |||||
|
MORN2_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 17) | NC score | 0.500892 (rank : 15) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6UL01 | Gene names | Morn2, Mopt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 2 (MORN motif protein in testis). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 18) | NC score | 0.073101 (rank : 30) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 19) | NC score | 0.061317 (rank : 38) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ANKY1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 20) | NC score | 0.218796 (rank : 22) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P2S6, Q8IYX5, Q8NDK5, Q9NX10 | Gene names | ANKMY1, TSAL1, ZMYND13 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 1 (Testis-specific ankyrin-like protein 1) (Zinc-finger MYND domain-containing protein 13). | |||||
|
EMIL2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 21) | NC score | 0.041351 (rank : 61) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BXX0, Q8NBH3, Q96JQ4 | Gene names | EMILIN2 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-2 precursor (Elastin microfibril interface-located protein 2) (Elastin microfibril interfacer 2) (Protein FOAP-10). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 22) | NC score | 0.039518 (rank : 64) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
DHX9_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 23) | NC score | 0.064569 (rank : 34) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 24) | NC score | 0.052889 (rank : 49) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
DMRTD_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 25) | NC score | 0.039639 (rank : 63) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IXT2, Q8N6Q2, Q96M39, Q96SD4 | Gene names | DMRTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2. | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.163984 (rank : 26) | NC score | 0.032703 (rank : 72) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 27) | NC score | 0.054131 (rank : 46) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.023485 (rank : 94) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
AATK_MOUSE
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.006900 (rank : 124) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80YE4, O35211, Q3U2U5, Q3UHR8, Q66JN3, Q80YE3, Q8CB63, Q8CHE2 | Gene names | Aatk, Kiaa0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 0.279714 (rank : 30) | NC score | 0.059282 (rank : 40) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
HORN_MOUSE
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.090603 (rank : 28) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
HRX_HUMAN
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.040644 (rank : 62) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.033655 (rank : 69) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.038061 (rank : 66) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SETD7_HUMAN
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.233074 (rank : 20) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WTS6, Q9C0E6 | Gene names | SETD7, KIAA1717, SET7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7) (Set9) (SET7/9). | |||||
|
SETD7_MOUSE
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.231488 (rank : 21) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VHL1 | Gene names | Setd7, Set7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.058077 (rank : 41) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
UBP16_HUMAN
|
||||||
| θ value | 0.365318 (rank : 38) | NC score | 0.018983 (rank : 103) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
HORN_HUMAN
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.102599 (rank : 27) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
INVS_MOUSE
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.013479 (rank : 113) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.050887 (rank : 55) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
UB7I1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.024996 (rank : 90) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
H11_MOUSE
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.028466 (rank : 79) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P43275 | Gene names | Hist1h1a, H1f1 | |||
|
Domain Architecture |
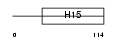
|
|||||
| Description | Histone H1.1 (H1 VAR.3) (H1a). | |||||
|
K1C10_HUMAN
|
||||||
| θ value | 0.62314 (rank : 44) | NC score | 0.027783 (rank : 83) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P13645, Q14664, Q8N175 | Gene names | KRT10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10). | |||||
|
BACH2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.010306 (rank : 119) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYV9, Q59H70, Q5T793, Q9NTS5 | Gene names | BACH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.036015 (rank : 68) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
MYPN_MOUSE
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.011079 (rank : 116) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5DTJ9, Q7TPW5, Q8BZ76 | Gene names | Mypn, Kiaa4170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myopalladin. | |||||
|
FA98B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.070060 (rank : 32) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80VD1, Q9CS28, Q9CS37 | Gene names | Fam98b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM98B. | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.044035 (rank : 60) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.055295 (rank : 43) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SAMH1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.024277 (rank : 92) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y3Z3, Q5JXG8, Q9H004, Q9H005, Q9H3U9 | Gene names | SAMHD1, MOP5 | |||
|
Domain Architecture |
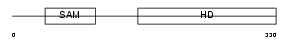
|
|||||
| Description | SAM domain and HD domain-containing protein 1 (Dendritic cell-derived IFNG-induced protein) (DCIP) (Monocyte protein 5) (MOP-5). | |||||
|
CD2L7_HUMAN
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.005052 (rank : 128) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.032402 (rank : 74) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
LORI_MOUSE
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.238791 (rank : 19) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
LTBP4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.007897 (rank : 122) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.053398 (rank : 47) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
SASH1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.027854 (rank : 82) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O94885, Q8TAI0, Q9H7R7 | Gene names | SASH1, KIAA0790, PEPE1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM and SH3 domain-containing protein 1 (Proline-glutamate repeat- containing protein). | |||||
|
SASH1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.027928 (rank : 81) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P59808 | Gene names | Sash1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM and SH3 domain-containing protein 1. | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.067702 (rank : 33) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
STRC_MOUSE
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.024189 (rank : 93) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VIM6 | Gene names | Strc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stereocilin precursor. | |||||
|
DHX9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.044244 (rank : 59) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
FA98A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.120191 (rank : 25) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NCA5 | Gene names | FAM98A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM98A. | |||||
|
TARA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.038382 (rank : 65) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
LEUK_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.029067 (rank : 77) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P15702 | Gene names | Spn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Ly- 48) (B cell differentiation antigen LP-3) (CD43 antigen). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.037920 (rank : 67) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.016252 (rank : 108) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
SYNPO_MOUSE
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.028916 (rank : 78) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.024703 (rank : 91) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CCD96_MOUSE
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.017926 (rank : 104) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
FA98A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.114320 (rank : 26) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3TJZ6, Q99KJ2 | Gene names | Fam98a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM98A. | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.013423 (rank : 114) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
NRIP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.015669 (rank : 110) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48552, Q8IWE8 | Gene names | NRIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-interacting protein 1 (Nuclear factor RIP140) (Receptor-interacting protein 140). | |||||
|
OGFR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.051740 (rank : 53) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
SOS1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.019406 (rank : 100) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.073542 (rank : 29) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SYNPO_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.032043 (rank : 75) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
TAB2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.013651 (rank : 112) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
TBCD2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.019306 (rank : 102) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
TCF20_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.027238 (rank : 84) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
ATX7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.023032 (rank : 96) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
CLCN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.006919 (rank : 123) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35523 | Gene names | CLCN1, CLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Chloride channel protein, skeletal muscle (Chloride channel protein 1) (ClC-1). | |||||
|
EMIL2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.025437 (rank : 88) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K482 | Gene names | Emilin2 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-2 precursor (Elastin microfibril interface-located protein 2) (Elastin microfibril interfacer 2) (Basilin). | |||||
|
KCNH3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.012519 (rank : 115) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.032673 (rank : 73) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.028387 (rank : 80) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.006410 (rank : 126) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
RBP56_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.071074 (rank : 31) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92804, Q92751 | Gene names | TAF15, RBP56, TAF2N | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 2N (RNA-binding protein 56) (TAFII68) (TAF(II)68). | |||||
|
SC6A5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.004307 (rank : 130) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q761V0, Q8CFM5, Q91ZQ2 | Gene names | Slc6a5, Glyt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent glycine transporter 2 (GlyT2) (GlyT-2) (Solute carrier family 6 member 5). | |||||
|
ACD_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.019329 (rank : 101) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96AP0, Q562H5, Q9H8F9 | Gene names | ACD, PIP1, PTOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adrenocortical dysplasia protein homolog (POT1 and TIN2-interacting protein). | |||||
|
ARSE_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.016596 (rank : 106) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51690, Q53FT2, Q53FU8 | Gene names | ARSE | |||
|
Domain Architecture |
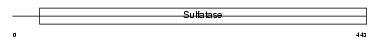
|
|||||
| Description | Arylsulfatase E precursor (EC 3.1.6.-) (ASE). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.050364 (rank : 57) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SOS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.020864 (rank : 98) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.026757 (rank : 85) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
ZN473_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | -0.002711 (rank : 134) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 824 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BI67, Q8BI98, Q8BIB7 | Gene names | Znf473, Zfp100, Zfp473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 473 homolog (Zinc finger protein 100) (Zfp-100). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.031760 (rank : 76) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
GP158_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.017127 (rank : 105) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.025776 (rank : 87) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.033214 (rank : 70) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.049196 (rank : 58) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PAK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | -0.001321 (rank : 132) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 878 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88643 | Gene names | Pak1, Paka | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 1 (EC 2.7.11.1) (p21-activated kinase 1) (PAK-1) (P65-PAK) (Alpha-PAK) (CDC42/RAC effector kinase PAK-A). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.026647 (rank : 86) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
APBB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.006752 (rank : 125) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.023389 (rank : 95) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
BAT3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.020486 (rank : 99) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
CAC1F_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.002345 (rank : 131) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CREB5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.008341 (rank : 120) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.015900 (rank : 109) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
ENAM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.014167 (rank : 111) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
GP108_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.007898 (rank : 121) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91WD0, Q8BXE9, Q925B1 | Gene names | Gpr108, Lustr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein GPR108 precursor (Lung seven transmembrane receptor 2). | |||||
|
L2GL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.004732 (rank : 129) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15334, O00188, Q58F11, Q86UK6 | Gene names | LLGL1, HUGL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(2) giant larvae protein homolog 1 (LLGL) (Human homolog to the D-lgl gene) (Hugl-1) (DLG4). | |||||
|
MUCDL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.010820 (rank : 117) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
MYCB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.032724 (rank : 71) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
PAK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | -0.001660 (rank : 133) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13153, O75561, Q13567, Q32M53, Q32M54, Q86W79 | Gene names | PAK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 1 (EC 2.7.11.1) (p21-activated kinase 1) (PAK-1) (P65-PAK) (Alpha-PAK). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.054780 (rank : 45) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
PTTG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.010724 (rank : 118) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQJ7, O88887, Q9Z2E6 | Gene names | Pttg1, Pttg | |||
|
Domain Architecture |

|
|||||
| Description | Securin (Pituitary tumor-transforming protein 1). | |||||
|
RTP3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.021633 (rank : 97) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
SIN3B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.005996 (rank : 127) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75182, Q68GC2, Q6P4B8, Q8TB34, Q9BSC8 | Gene names | SIN3B, KIAA0700 | |||
|
Domain Architecture |
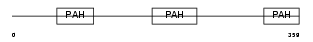
|
|||||
| Description | Paired amphipathic helix protein Sin3b (Transcriptional corepressor Sin3b) (Histone deacetylase complex subunit Sin3b). | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.016292 (rank : 107) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
ZCPW1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.025101 (rank : 89) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
ZN690_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | -0.003012 (rank : 135) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IWY8, Q32M75, Q32M76, Q8NA40 | Gene names | ZNF690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 690. | |||||
|
BAT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.053160 (rank : 48) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
KR193_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.148483 (rank : 24) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z4W3 | Gene names | KRTAP19-3, KAP19.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-3 (Glycine/tyrosine-rich protein) (GTHRP). | |||||
|
KR197_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.063578 (rank : 36) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3SYF9 | Gene names | KRTAP19-7, KAP19.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-7. | |||||
|
KR202_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.182528 (rank : 23) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3LI61 | Gene names | KRTAP20-2, KAP20.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 20-2. | |||||
|
LORI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.244206 (rank : 18) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.052655 (rank : 50) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.055069 (rank : 44) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.061491 (rank : 37) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.051794 (rank : 52) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.061135 (rank : 39) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
PRPE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.057297 (rank : 42) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P02811 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide P-E (IB-9). | |||||
|
ROA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.051732 (rank : 54) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P09651, Q6PJZ7 | Gene names | HNRPA1 | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A1 (Helix-destabilizing protein) (Single-strand RNA-binding protein) (hnRNP core protein A1). | |||||
|
TB182_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.052293 (rank : 51) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
TMAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.064381 (rank : 35) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
TTDN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.050748 (rank : 56) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TAP9 | Gene names | TTDN1, C7orf11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TTD non-photosensitive 1 protein. | |||||
|
JPH1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 120 | |
| SwissProt Accessions | Q9ET80, Q9EQZ3 | Gene names | Jph1, Jp1 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
JPH1_HUMAN
|
||||||
| NC score | 0.987102 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9HDC5 | Gene names | JPH1, JP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
JPH3_HUMAN
|
||||||
| NC score | 0.968144 (rank : 3) | θ value | 1.20145e-153 (rank : 5) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WXH2, Q8N471, Q9HDC3, Q9HDC4 | Gene names | JPH3, JP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
JPH3_MOUSE
|
||||||
| NC score | 0.967876 (rank : 4) | θ value | 4.56552e-153 (rank : 6) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ET77, Q9EQZ2 | Gene names | Jph3, Jp3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
JPH4_HUMAN
|
||||||
| NC score | 0.946048 (rank : 5) | θ value | 2.46047e-98 (rank : 8) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96JJ6, Q8ND44, Q96DQ0 | Gene names | JPH4, JPHL1, KIAA1831 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
JPH4_MOUSE
|
||||||
| NC score | 0.944674 (rank : 6) | θ value | 4.95767e-99 (rank : 7) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q80WT0, Q8BMI1 | Gene names | Jph4, Jphl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
JPH2_MOUSE
|
||||||
| NC score | 0.936563 (rank : 7) | θ value | 6.37065e-155 (rank : 4) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.877916 (rank : 8) | θ value | 1.24043e-158 (rank : 3) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
MORN3_MOUSE
|
||||||
| NC score | 0.673943 (rank : 9) | θ value | 5.43371e-13 (rank : 9) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8C5T4, Q8VE21, Q9D5H6 | Gene names | Morn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 3. | |||||
|
MORN3_HUMAN
|
||||||
| NC score | 0.665912 (rank : 10) | θ value | 9.26847e-13 (rank : 10) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PF18, Q86YQ9 | Gene names | MORN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 3. | |||||
|
TSGA2_HUMAN
|
||||||
| NC score | 0.652526 (rank : 11) | θ value | 7.84624e-12 (rank : 12) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WYR4 | Gene names | TSGA2, TSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific gene A2 protein (Male meiotic metaphase chromosome- associated acidic protein) (Meichroacidin). | |||||
|
TSGA2_MOUSE
|
||||||
| NC score | 0.645466 (rank : 12) | θ value | 6.00763e-12 (rank : 11) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VIG3, Q9DAL5 | Gene names | Tsga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific gene A2 protein (Male meiotic metaphase chromosome- associated acidic protein) (Meichroacidin). | |||||
|
MORN1_HUMAN
|
||||||
| NC score | 0.622742 (rank : 13) | θ value | 6.64225e-11 (rank : 13) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5T089, Q8WW30, Q9H852 | Gene names | MORN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 1. | |||||
|
MORN2_HUMAN
|
||||||
| NC score | 0.517877 (rank : 14) | θ value | 0.00035302 (rank : 16) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q502X0, Q6UL00 | Gene names | MORN2, MOPT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 2 (MORN motif protein in testis). | |||||
|
MORN2_MOUSE
|
||||||
| NC score | 0.500892 (rank : 15) | θ value | 0.000602161 (rank : 17) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6UL01 | Gene names | Morn2, Mopt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 2 (MORN motif protein in testis). | |||||
|
ALS2_HUMAN
|
||||||
| NC score | 0.413604 (rank : 16) | θ value | 1.9326e-10 (rank : 14) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
ALS2_MOUSE
|
||||||
| NC score | 0.413582 (rank : 17) | θ value | 1.9326e-10 (rank : 15) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q920R0, Q8JZR1, Q9CXJ3 | Gene names | Als2 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2 homolog). | |||||
|
LORI_HUMAN
|
||||||
| NC score | 0.244206 (rank : 18) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
LORI_MOUSE
|
||||||
| NC score | 0.238791 (rank : 19) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
SETD7_HUMAN
|
||||||
| NC score | 0.233074 (rank : 20) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WTS6, Q9C0E6 | Gene names | SETD7, KIAA1717, SET7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7) (Set9) (SET7/9). | |||||
|
SETD7_MOUSE
|
||||||
| NC score | 0.231488 (rank : 21) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VHL1 | Gene names | Setd7, Set7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7). | |||||
|
ANKY1_HUMAN
|
||||||
| NC score | 0.218796 (rank : 22) | θ value | 0.0431538 (rank : 20) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P2S6, Q8IYX5, Q8NDK5, Q9NX10 | Gene names | ANKMY1, TSAL1, ZMYND13 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 1 (Testis-specific ankyrin-like protein 1) (Zinc-finger MYND domain-containing protein 13). | |||||
|
KR202_HUMAN
|
||||||
| NC score | 0.182528 (rank : 23) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3LI61 | Gene names | KRTAP20-2, KAP20.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 20-2. | |||||
|
KR193_HUMAN
|
||||||
| NC score | 0.148483 (rank : 24) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z4W3 | Gene names | KRTAP19-3, KAP19.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-3 (Glycine/tyrosine-rich protein) (GTHRP). | |||||
|
FA98A_HUMAN
|
||||||
| NC score | 0.120191 (rank : 25) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NCA5 | Gene names | FAM98A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM98A. | |||||
|
FA98A_MOUSE
|
||||||
| NC score | 0.114320 (rank : 26) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3TJZ6, Q99KJ2 | Gene names | Fam98a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM98A. | |||||
|
HORN_HUMAN
|
||||||
| NC score | 0.102599 (rank : 27) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
HORN_MOUSE
|
||||||
| NC score | 0.090603 (rank : 28) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.073542 (rank : 29) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.073101 (rank : 30) | θ value | 0.00134147 (rank : 18) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
RBP56_HUMAN
|
||||||
| NC score | 0.071074 (rank : 31) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92804, Q92751 | Gene names | TAF15, RBP56, TAF2N | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 2N (RNA-binding protein 56) (TAFII68) (TAF(II)68). | |||||
|
FA98B_MOUSE
|
||||||
| NC score | 0.070060 (rank : 32) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80VD1, Q9CS28, Q9CS37 | Gene names | Fam98b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM98B. | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.067702 (rank : 33) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
DHX9_MOUSE
|
||||||
| NC score | 0.064569 (rank : 34) | θ value | 0.0736092 (rank : 23) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
TMAP1_HUMAN
|
||||||
| NC score | 0.064381 (rank : 35) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
KR197_HUMAN
|
||||||
| NC score | 0.063578 (rank : 36) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3SYF9 | Gene names | KRTAP19-7, KAP19.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-7. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.061491 (rank : 37) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.061317 (rank : 38) | θ value | 0.00509761 (rank : 19) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PRP5_HUMAN
|
||||||
| NC score | 0.061135 (rank : 39) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.059282 (rank : 40) | θ value | 0.279714 (rank : 30) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.058077 (rank : 41) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
PRPE_HUMAN
|
||||||
| NC score | 0.057297 (rank : 42) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P02811 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide P-E (IB-9). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.055295 (rank : 43) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.055069 (rank : 44) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.054780 (rank : 45) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.054131 (rank : 46) | θ value | 0.21417 (rank : 27) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.053398 (rank : 47) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.053160 (rank : 48) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.052889 (rank : 49) | θ value | 0.0736092 (rank : 24) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.052655 (rank : 50) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
TB182_HUMAN
|
||||||
| NC score | 0.052293 (rank : 51) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.051794 (rank : 52) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.051740 (rank : 53) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
ROA1_HUMAN
|
||||||
| NC score | 0.051732 (rank : 54) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P09651, Q6PJZ7 | Gene names | HNRPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A1 (Helix-destabilizing protein) (Single-strand RNA-binding protein) (hnRNP core protein A1). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.050887 (rank : 55) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
TTDN1_HUMAN
|
||||||
| NC score | 0.050748 (rank : 56) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TAP9 | Gene names | TTDN1, C7orf11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TTD non-photosensitive 1 protein. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.050364 (rank : 57) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.049196 (rank : 58) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
DHX9_HUMAN
|
||||||
| NC score | 0.044244 (rank : 59) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.044035 (rank : 60) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
EMIL2_HUMAN
|
||||||
| NC score | 0.041351 (rank : 61) | θ value | 0.0563607 (rank : 21) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BXX0, Q8NBH3, Q96JQ4 | Gene names | EMILIN2 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-2 precursor (Elastin microfibril interface-located protein 2) (Elastin microfibril interfacer 2) (Protein FOAP-10). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.040644 (rank : 62) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
DMRTD_HUMAN
|
||||||
| NC score | 0.039639 (rank : 63) | θ value | 0.0961366 (rank : 25) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IXT2, Q8N6Q2, Q96M39, Q96SD4 | Gene names | DMRTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2. | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.039518 (rank : 64) | θ value | 0.0563607 (rank : 22) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.038382 (rank : 65) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.038061 (rank : 66) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.037920 (rank : 67) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.036015 (rank : 68) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.033655 (rank : 69) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.033214 (rank : 70) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
MYCB2_HUMAN
|
||||||
| NC score | 0.032724 (rank : 71) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.032703 (rank : 72) | θ value | 0.163984 (rank : 26) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.032673 (rank : 73) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.032402 (rank : 74) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
SYNPO_HUMAN
|
||||||
| NC score | 0.032043 (rank : 75) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.031760 (rank : 76) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
LEUK_MOUSE
|
||||||
| NC score | 0.029067 (rank : 77) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P15702 | Gene names | Spn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Ly- 48) (B cell differentiation antigen LP-3) (CD43 antigen). | |||||
|
SYNPO_MOUSE
|
||||||
| NC score | 0.028916 (rank : 78) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
H11_MOUSE
|
||||||
| NC score | 0.028466 (rank : 79) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P43275 | Gene names | Hist1h1a, H1f1 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.1 (H1 VAR.3) (H1a). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.028387 (rank : 80) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
SASH1_MOUSE
|
||||||
| NC score | 0.027928 (rank : 81) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P59808 | Gene names | Sash1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM and SH3 domain-containing protein 1. | |||||
|
SASH1_HUMAN
|
||||||
| NC score | 0.027854 (rank : 82) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O94885, Q8TAI0, Q9H7R7 | Gene names | SASH1, KIAA0790, PEPE1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM and SH3 domain-containing protein 1 (Proline-glutamate repeat- containing protein). | |||||
|
K1C10_HUMAN
|
||||||
| NC score | 0.027783 (rank : 83) | θ value | 0.62314 (rank : 44) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P13645, Q14664, Q8N175 | Gene names | KRT10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10). | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.027238 (rank : 84) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.026757 (rank : 85) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.026647 (rank : 86) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.025776 (rank : 87) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
EMIL2_MOUSE
|
||||||
| NC score | 0.025437 (rank : 88) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K482 | Gene names | Emilin2 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-2 precursor (Elastin microfibril interface-located protein 2) (Elastin microfibril interfacer 2) (Basilin). | |||||
|
ZCPW1_MOUSE
|
||||||
| NC score | 0.025101 (rank : 89) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
UB7I1_MOUSE
|
||||||
| NC score | 0.024996 (rank : 90) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.024703 (rank : 91) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
SAMH1_HUMAN
|
||||||
| NC score | 0.024277 (rank : 92) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y3Z3, Q5JXG8, Q9H004, Q9H005, Q9H3U9 | Gene names | SAMHD1, MOP5 | |||
|
Domain Architecture |

|
|||||
| Description | SAM domain and HD domain-containing protein 1 (Dendritic cell-derived IFNG-induced protein) (DCIP) (Monocyte protein 5) (MOP-5). | |||||
|
STRC_MOUSE
|
||||||
| NC score | 0.024189 (rank : 93) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VIM6 | Gene names | Strc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stereocilin precursor. | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.023485 (rank : 94) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.023389 (rank : 95) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
ATX7_HUMAN
|
||||||
| NC score | 0.023032 (rank : 96) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
RTP3_MOUSE
|
||||||
| NC score | 0.021633 (rank : 97) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
SOS1_MOUSE
|
||||||
| NC score | 0.020864 (rank : 98) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
BAT3_MOUSE
|
||||||
| NC score | 0.020486 (rank : 99) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
SOS1_HUMAN
|
||||||
| NC score | 0.019406 (rank : 100) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
ACD_HUMAN
|
||||||
| NC score | 0.019329 (rank : 101) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96AP0, Q562H5, Q9H8F9 | Gene names | ACD, PIP1, PTOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adrenocortical dysplasia protein homolog (POT1 and TIN2-interacting protein). | |||||
|
TBCD2_HUMAN
|
||||||
| NC score | 0.019306 (rank : 102) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
UBP16_HUMAN
|
||||||
| NC score | 0.018983 (rank : 103) | θ value | 0.365318 (rank : 38) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
CCD96_MOUSE
|
||||||
| NC score | 0.017926 (rank : 104) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
GP158_HUMAN
|
||||||
| NC score | 0.017127 (rank : 105) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
ARSE_HUMAN
|
||||||
| NC score | 0.016596 (rank : 106) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51690, Q53FT2, Q53FU8 | Gene names | ARSE | |||
|
Domain Architecture |

|
|||||
| Description | Arylsulfatase E precursor (EC 3.1.6.-) (ASE). | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.016292 (rank : 107) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.016252 (rank : 108) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.015900 (rank : 109) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
NRIP1_HUMAN
|
||||||
| NC score | 0.015669 (rank : 110) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48552, Q8IWE8 | Gene names | NRIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-interacting protein 1 (Nuclear factor RIP140) (Receptor-interacting protein 140). | |||||
|
ENAM_HUMAN
|
||||||
| NC score | 0.014167 (rank : 111) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
TAB2_MOUSE
|
||||||
| NC score | 0.013651 (rank : 112) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
INVS_MOUSE
|
||||||
| NC score | 0.013479 (rank : 113) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.013423 (rank : 114) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
KCNH3_HUMAN
|
||||||
| NC score | 0.012519 (rank : 115) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
MYPN_MOUSE
|
||||||
| NC score | 0.011079 (rank : 116) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5DTJ9, Q7TPW5, Q8BZ76 | Gene names | Mypn, Kiaa4170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myopalladin. | |||||
|
MUCDL_MOUSE
|
||||||
| NC score | 0.010820 (rank : 117) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
PTTG1_MOUSE
|
||||||
| NC score | 0.010724 (rank : 118) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQJ7, O88887, Q9Z2E6 | Gene names | Pttg1, Pttg | |||
|
Domain Architecture |

|
|||||
| Description | Securin (Pituitary tumor-transforming protein 1). | |||||
|
BACH2_HUMAN
|
||||||
| NC score | 0.010306 (rank : 119) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYV9, Q59H70, Q5T793, Q9NTS5 | Gene names | BACH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
CREB5_HUMAN
|
||||||
| NC score | 0.008341 (rank : 120) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
GP108_MOUSE
|
||||||
| NC score | 0.007898 (rank : 121) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91WD0, Q8BXE9, Q925B1 | Gene names | Gpr108, Lustr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein GPR108 precursor (Lung seven transmembrane receptor 2). | |||||
|
LTBP4_MOUSE
|
||||||
| NC score | 0.007897 (rank : 122) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
CLCN1_HUMAN
|
||||||
| NC score | 0.006919 (rank : 123) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35523 | Gene names | CLCN1, CLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Chloride channel protein, skeletal muscle (Chloride channel protein 1) (ClC-1). | |||||
|
AATK_MOUSE
|
||||||
| NC score | 0.006900 (rank : 124) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80YE4, O35211, Q3U2U5, Q3UHR8, Q66JN3, Q80YE3, Q8CB63, Q8CHE2 | Gene names | Aatk, Kiaa0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase). | |||||
|
APBB2_MOUSE
|
||||||
| NC score | 0.006752 (rank : 125) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.006410 (rank : 126) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
SIN3B_HUMAN
|
||||||
| NC score | 0.005996 (rank : 127) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75182, Q68GC2, Q6P4B8, Q8TB34, Q9BSC8 | Gene names | SIN3B, KIAA0700 | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3b (Transcriptional corepressor Sin3b) (Histone deacetylase complex subunit Sin3b). | |||||
|
CD2L7_HUMAN
|
||||||
| NC score | 0.005052 (rank : 128) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
L2GL1_HUMAN
|
||||||
| NC score | 0.004732 (rank : 129) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15334, O00188, Q58F11, Q86UK6 | Gene names | LLGL1, HUGL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(2) giant larvae protein homolog 1 (LLGL) (Human homolog to the D-lgl gene) (Hugl-1) (DLG4). | |||||
|
SC6A5_MOUSE
|
||||||
| NC score | 0.004307 (rank : 130) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q761V0, Q8CFM5, Q91ZQ2 | Gene names | Slc6a5, Glyt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent glycine transporter 2 (GlyT2) (GlyT-2) (Solute carrier family 6 member 5). | |||||
|
CAC1F_HUMAN
|
||||||
| NC score | 0.002345 (rank : 131) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
PAK1_MOUSE
|
||||||
| NC score | -0.001321 (rank : 132) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 878 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88643 | Gene names | Pak1, Paka | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 1 (EC 2.7.11.1) (p21-activated kinase 1) (PAK-1) (P65-PAK) (Alpha-PAK) (CDC42/RAC effector kinase PAK-A). | |||||
|
PAK1_HUMAN
|
||||||
| NC score | -0.001660 (rank : 133) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13153, O75561, Q13567, Q32M53, Q32M54, Q86W79 | Gene names | PAK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 1 (EC 2.7.11.1) (p21-activated kinase 1) (PAK-1) (P65-PAK) (Alpha-PAK). | |||||
|
ZN473_MOUSE
|
||||||
| NC score | -0.002711 (rank : 134) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 824 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BI67, Q8BI98, Q8BIB7 | Gene names | Znf473, Zfp100, Zfp473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 473 homolog (Zinc finger protein 100) (Zfp-100). | |||||
|
ZN690_HUMAN
|
||||||
| NC score | -0.003012 (rank : 135) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 120 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IWY8, Q32M75, Q32M76, Q8NA40 | Gene names | ZNF690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 690. | |||||