Please be patient as the page loads
|
KCNH3_HUMAN
|
||||||
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
KCNH3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 148 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
KCNH3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994671 (rank : 2) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
KCNH4_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.980859 (rank : 4) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
KCNH8_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.984717 (rank : 3) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96L42 | Gene names | KCNH8 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 8 (Voltage-gated potassium channel subunit Kv12.1) (Ether-a-go-go-like potassium channel 3) (ELK channel 3) (ELK3) (ELK1) (hElk1). | |||||
|
KCNH6_HUMAN
|
||||||
| θ value | 2.77624e-142 (rank : 5) | NC score | 0.960920 (rank : 9) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
KCNH1_MOUSE
|
||||||
| θ value | 2.35568e-133 (rank : 6) | NC score | 0.961927 (rank : 5) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
KCNH5_MOUSE
|
||||||
| θ value | 2.35568e-133 (rank : 7) | NC score | 0.961581 (rank : 8) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
KCNH5_HUMAN
|
||||||
| θ value | 4.01817e-133 (rank : 8) | NC score | 0.961603 (rank : 7) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
KCNH1_HUMAN
|
||||||
| θ value | 5.24793e-133 (rank : 9) | NC score | 0.961732 (rank : 6) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
KCNH2_MOUSE
|
||||||
| θ value | 1.02895e-112 (rank : 10) | NC score | 0.951699 (rank : 11) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
KCNH2_HUMAN
|
||||||
| θ value | 2.9938e-112 (rank : 11) | NC score | 0.951879 (rank : 10) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
KCNH7_MOUSE
|
||||||
| θ value | 2.89973e-107 (rank : 12) | NC score | 0.950001 (rank : 12) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
KCNH7_HUMAN
|
||||||
| θ value | 3.54468e-105 (rank : 13) | NC score | 0.948978 (rank : 13) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
CNGA3_HUMAN
|
||||||
| θ value | 8.91499e-32 (rank : 14) | NC score | 0.793382 (rank : 18) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16281, Q9UP64 | Gene names | CNGA3, CNCG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA1_MOUSE
|
||||||
| θ value | 1.52067e-31 (rank : 15) | NC score | 0.793801 (rank : 17) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_MOUSE
|
||||||
| θ value | 4.42448e-31 (rank : 16) | NC score | 0.797070 (rank : 16) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JJZ8, Q9WV01 | Gene names | Cnga3, Cng3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA2_MOUSE
|
||||||
| θ value | 1.28732e-30 (rank : 17) | NC score | 0.806607 (rank : 14) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q62398 | Gene names | Cnga2, Cncg2, Cncg4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated olfactory channel (Cyclic nucleotide-gated cation channel 2) (CNG channel 2) (CNG-2) (CNG2). | |||||
|
CNGA1_HUMAN
|
||||||
| θ value | 1.6813e-30 (rank : 18) | NC score | 0.797659 (rank : 15) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 1.4233e-29 (rank : 19) | NC score | 0.765714 (rank : 24) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
HCN1_HUMAN
|
||||||
| θ value | 2.42779e-29 (rank : 20) | NC score | 0.781330 (rank : 19) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
HCN1_MOUSE
|
||||||
| θ value | 2.42779e-29 (rank : 21) | NC score | 0.777668 (rank : 20) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
HCN2_MOUSE
|
||||||
| θ value | 5.40856e-29 (rank : 22) | NC score | 0.767830 (rank : 23) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 5.40856e-29 (rank : 23) | NC score | 0.764949 (rank : 25) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 7.06379e-29 (rank : 24) | NC score | 0.761466 (rank : 26) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
HCN3_MOUSE
|
||||||
| θ value | 6.61148e-27 (rank : 25) | NC score | 0.775874 (rank : 22) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
HCN3_HUMAN
|
||||||
| θ value | 1.12775e-26 (rank : 26) | NC score | 0.776507 (rank : 21) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
CNGB1_HUMAN
|
||||||
| θ value | 3.63628e-17 (rank : 27) | NC score | 0.715553 (rank : 29) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CNGB3_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 28) | NC score | 0.717019 (rank : 28) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CNGB3_MOUSE
|
||||||
| θ value | 4.91457e-14 (rank : 29) | NC score | 0.727910 (rank : 27) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JJZ9 | Gene names | Cngb3, Cng6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit) (Cyclic nucleotide-gated channel subunit CNG6). | |||||
|
KCNKH_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 30) | NC score | 0.249220 (rank : 30) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
KCNK3_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 31) | NC score | 0.178806 (rank : 35) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
KCNK3_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 32) | NC score | 0.182208 (rank : 32) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNKF_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 33) | NC score | 0.189068 (rank : 31) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
KCNKA_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 34) | NC score | 0.174029 (rank : 36) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
CCG8_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 35) | NC score | 0.019364 (rank : 98) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHW2 | Gene names | Cacng8 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-8 subunit (Neuronal voltage- gated calcium channel gamma-8 subunit). | |||||
|
KCNK2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 36) | NC score | 0.182061 (rank : 33) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
KCNK2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 37) | NC score | 0.181386 (rank : 34) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
MAST2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 38) | NC score | 0.004396 (rank : 146) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 39) | NC score | 0.034663 (rank : 71) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
BICD1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 40) | NC score | 0.015975 (rank : 113) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
TARA_MOUSE
|
||||||
| θ value | 0.163984 (rank : 41) | NC score | 0.017361 (rank : 105) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
KCNK4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 42) | NC score | 0.166641 (rank : 38) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
KGP1A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 43) | NC score | 0.042800 (rank : 63) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13976 | Gene names | PRKG1, PRKGR1A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, alpha isozyme (EC 2.7.11.12) (CGK 1 alpha) (cGKI-alpha). | |||||
|
KGP1B_HUMAN
|
||||||
| θ value | 0.21417 (rank : 44) | NC score | 0.042853 (rank : 62) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 921 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P14619 | Gene names | PRKG1, PRKG1B, PRKGR1B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.279714 (rank : 45) | NC score | 0.034905 (rank : 70) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
KCNK9_HUMAN
|
||||||
| θ value | 0.279714 (rank : 46) | NC score | 0.153035 (rank : 41) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NPC2, Q2M290, Q540F2 | Gene names | KCNK9, TASK3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3) (Two pore potassium channel KT3.2). | |||||
|
KCNK9_MOUSE
|
||||||
| θ value | 0.279714 (rank : 47) | NC score | 0.153184 (rank : 40) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q3LS21 | Gene names | Kcnk9, Task3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 48) | NC score | 0.020424 (rank : 93) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 49) | NC score | 0.016928 (rank : 110) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 50) | NC score | 0.017805 (rank : 104) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | 0.365318 (rank : 51) | NC score | 0.018233 (rank : 103) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 52) | NC score | 0.027013 (rank : 80) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 0.365318 (rank : 53) | NC score | 0.040390 (rank : 65) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
EPN3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 54) | NC score | 0.035038 (rank : 69) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H201, Q9BVN6, Q9NWK2 | Gene names | EPN3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
KCNK6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 55) | NC score | 0.171806 (rank : 37) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
KGP1B_MOUSE
|
||||||
| θ value | 0.47712 (rank : 56) | NC score | 0.042375 (rank : 64) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Z0Z0 | Gene names | Prkg1, Prkg1b, Prkgr1b | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
TS13_MOUSE
|
||||||
| θ value | 0.47712 (rank : 57) | NC score | 0.032954 (rank : 74) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01755 | Gene names | Tcp11 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific protein PBS13. | |||||
|
BICD1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 58) | NC score | 0.012589 (rank : 122) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 59) | NC score | 0.032166 (rank : 75) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
KCNK5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.125483 (rank : 45) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O95279 | Gene names | KCNK5, TASK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 5 (Acid-sensitive potassium channel protein TASK-2) (TWIK-related acid-sensitive K(+) channel 2). | |||||
|
RTP3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 61) | NC score | 0.021772 (rank : 88) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
SEM6B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 62) | NC score | 0.008367 (rank : 135) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H3T3, Q9NRK9 | Gene names | SEMA6B, SEMAZ | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin Z) (Sema Z). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 63) | NC score | 0.036709 (rank : 66) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SENP3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 64) | NC score | 0.014601 (rank : 118) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EP97 | Gene names | Senp3, Smt3ip, Smt3ip1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 3 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP3) (SUMO-1-specific protease 3) (Smt3-specific isopeptidase 1) (Smt3ip1). | |||||
|
KCNK4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.152361 (rank : 42) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
|
KCNKG_HUMAN
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.158686 (rank : 39) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
SON_HUMAN
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.017306 (rank : 106) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CMTA2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.016866 (rank : 111) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80Y50, Q3TFK0, Q5SX68, Q80TP1, Q8R0D9, Q8R2N5 | Gene names | Camta2, Kiaa0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
BIN1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.020321 (rank : 95) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08539, Q62434 | Gene names | Bin1, Amphl, Sh3p9 | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (SH3-domain-containing protein 9). | |||||
|
CEP68_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.025124 (rank : 82) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q76N32, O60326, Q9BQ18, Q9UDM9 | Gene names | CEP68, KIAA0582 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
DAB2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.018446 (rank : 101) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
KKCC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.000567 (rank : 153) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N5S9, Q9BQH3 | Gene names | CAMKK1, CAMKKA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase kinase 1 (EC 2.7.11.17) (Calcium/calmodulin-dependent protein kinase kinase alpha) (CaM-kinase kinase alpha) (CaM-KK alpha) (CaMKK alpha) (CaMKK 1) (CaM-kinase IV kinase). | |||||
|
LCOR_MOUSE
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.021764 (rank : 89) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZPI3, Q3U302, Q5CZW7, Q80VA8, Q8BGT2, Q8C9Q0 | Gene names | Lcor, Kiaa1795, Mlr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ligand-dependent corepressor (LCoR) (Mblk1-related protein 2). | |||||
|
LIMD1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.010196 (rank : 130) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
PHLB1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.023954 (rank : 84) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.036260 (rank : 67) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
BIN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.023220 (rank : 86) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.015473 (rank : 116) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CXXC6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.023436 (rank : 85) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
FOXN1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.013680 (rank : 121) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
HAIR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.026850 (rank : 81) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
KCNN4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.036259 (rank : 68) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15554 | Gene names | KCNN4, IK1, IKCA1, KCA4, SK4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1) (IKCa1) (Putative Gardos channel). | |||||
|
PDZD6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.017252 (rank : 108) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULD6, Q4W5I8, Q86V55 | Gene names | PDZD6, KIAA1284, PDZK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 6. | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.007406 (rank : 137) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
EGR4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | -0.002502 (rank : 158) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
FOXN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.017278 (rank : 107) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15353, O15352 | Gene names | FOXN1, WHN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude). | |||||
|
GA2L2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.012166 (rank : 124) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
INVS_MOUSE
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.006408 (rank : 142) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
KCNK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.126119 (rank : 43) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O00180, Q13307 | Gene names | KCNK1, HOHO1, KCNO1, TWIK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1) (Potassium channel KCNO1). | |||||
|
KCNN4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.033355 (rank : 73) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O89109, Q3U1J8, Q9CY39 | Gene names | Kcnn4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1). | |||||
|
LCOR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.019948 (rank : 97) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96JN0, Q5VW16, Q7Z723, Q86T32, Q86T33, Q8N3L6 | Gene names | LCOR, KIAA1795, MLR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ligand-dependent corepressor (LCoR) (Mblk1-related protein 2). | |||||
|
MINP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.032146 (rank : 76) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UNW1, O95172, O95286, Q59EJ2, Q9UGA3 | Gene names | MINPP1, MIPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple inositol polyphosphate phosphatase 1 precursor (EC 3.1.3.62) (Inositol (1,3,4,5)-tetrakisphosphate 3-phosphatase) (Ins(1,3,4,5)P(4) 3-phosphatase). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.018638 (rank : 100) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PK2L1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.010698 (rank : 128) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.043056 (rank : 61) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
PSL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.018282 (rank : 102) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3TD49, Q3TLQ8, Q3UG60, Q80VZ6 | Gene names | Sppl2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.030584 (rank : 78) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.031556 (rank : 77) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.006517 (rank : 141) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
XKR7_MOUSE
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.017194 (rank : 109) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5GH64 | Gene names | Xkr7, Xrg7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 7. | |||||
|
CK056_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.011023 (rank : 126) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N612, Q9C0A4, Q9H0N3, Q9H624 | Gene names | C11orf56, KIAA1759 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf56. | |||||
|
EPAS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.004394 (rank : 147) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
JPH1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.012519 (rank : 123) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ET80, Q9EQZ3 | Gene names | Jph1, Jp1 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
KCNK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.122328 (rank : 46) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O08581 | Gene names | Kcnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.007916 (rank : 136) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
NBEA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.008818 (rank : 134) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NFP9, Q9HCM8, Q9NSU1, Q9NW98, Q9Y6J1 | Gene names | NBEA, BCL8B, KIAA1544, LYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2) (Protein BCL8B). | |||||
|
NBEA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.008931 (rank : 133) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EPN1, Q8C931, Q9EPM9, Q9EPN0, Q9WVM9 | Gene names | Nbea, Lyst2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.001724 (rank : 151) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.011729 (rank : 125) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SUSD4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.006143 (rank : 143) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BH32, Q8VC43 | Gene names | Susd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
APXL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.010323 (rank : 129) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
CAMKV_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.003255 (rank : 150) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
CI079_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.020210 (rank : 96) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
CJ108_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.016114 (rank : 112) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N8Z3 | Gene names | C10orf108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative uncharacterized protein C10orf108. | |||||
|
DUS2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.005672 (rank : 144) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q05923 | Gene names | DUSP2, PAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
EPN2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.014473 (rank : 119) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
|
KCND1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.046992 (rank : 60) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KSR1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | -0.000022 (rank : 154) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61097, Q61648, Q78DX8 | Gene names | Ksr1, Ksr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras) (mKSR1) (Hb protein). | |||||
|
STK10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | -0.000650 (rank : 155) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
ZN703_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.014751 (rank : 117) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H7S9, Q5XG76 | Gene names | ZNF703 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 703. | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.007058 (rank : 139) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
KCNN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.021593 (rank : 90) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H2S1 | Gene names | KCNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.021570 (rank : 91) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P58390 | Gene names | Kcnn2, Sk2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
M3K10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | -0.000873 (rank : 156) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q02779, Q12761, Q14871 | Gene names | MAP3K10, MLK2, MST | |||
|
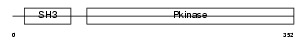
Domain Architecture |
|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 10 (EC 2.7.11.25) (Mixed lineage kinase 2) (Protein kinase MST). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.010160 (rank : 131) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.015517 (rank : 115) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.034308 (rank : 72) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.028458 (rank : 79) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.018868 (rank : 99) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
ZN227_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | -0.003956 (rank : 161) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 781 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86WZ6 | Gene names | ZNF227 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 227. | |||||
|
CKLF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.022903 (rank : 87) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBR5, Q9UHM7, Q9UHN8, Q9UI41 | Gene names | CKLF | |||
|
Domain Architecture |

|
|||||
| Description | Chemokine-like factor (C32). | |||||
|
DGKG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.001362 (rank : 152) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91WG7 | Gene names | Dgkg, Dagk3 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase gamma (EC 2.7.1.107) (Diglyceride kinase gamma) (DGK-gamma) (DAG kinase gamma) (88 kDa diacylglycerol kinase). | |||||
|
EMID1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.005194 (rank : 145) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91VF5 | Gene names | Emid1, Emu1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EMI domain-containing protein 1 precursor (Protein Emu1) (Emilin and multimerin domain-containing protein 1). | |||||
|
EPN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.020754 (rank : 92) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.024748 (rank : 83) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
HD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.009071 (rank : 132) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
IRS2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.015859 (rank : 114) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
JPH2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.013811 (rank : 120) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
K0179_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.010943 (rank : 127) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14684, Q8TBZ4 | Gene names | KIAA0179 | |||
|
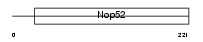
Domain Architecture |
|
|||||
| Description | Protein KIAA0179. | |||||
|
MTR1L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | -0.002633 (rank : 159) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1180 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13585 | Gene names | GPR50 | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin-related receptor (G protein-coupled receptor 50) (H9). | |||||
|
PANK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.003904 (rank : 148) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K4K6, Q9D3K1, Q9QXM8 | Gene names | Pank1, Pank | |||
|
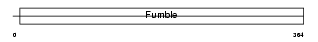
Domain Architecture |
|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (mPank1) (mPank). | |||||
|
PLXB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.006715 (rank : 140) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
PTC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.003762 (rank : 149) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6C5, O95341, O95856, Q5QP87, Q6UX14 | Gene names | PTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 2 (PTC2). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.020381 (rank : 94) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
VASN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | -0.001514 (rank : 157) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6EMK4, Q6UXL4, Q6UXL5, Q96CX1 | Gene names | VASN, SLITL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vasorin precursor (Protein Slit-like 2). | |||||
|
WD21C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.007335 (rank : 138) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NA75 | Gene names | WDR21C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 21C. | |||||
|
ZF276_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | -0.002805 (rank : 160) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N554 | Gene names | ZFP276 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 276 homolog (Zfp-276). | |||||
|
ZN169_HUMAN
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | -0.004091 (rank : 162) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14929, Q6PI28 | Gene names | ZNF169 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 169. | |||||
|
CT152_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.059874 (rank : 58) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D5U8, Q8R083 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152 homolog. | |||||
|
KAP0_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.126066 (rank : 44) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
KAP0_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.115004 (rank : 47) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
KAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.104206 (rank : 48) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.101834 (rank : 49) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.073723 (rank : 54) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.075813 (rank : 52) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12367 | Gene names | Prkar2a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.073902 (rank : 53) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.077118 (rank : 51) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KCNK7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.052615 (rank : 59) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y2U2, Q9Y2U3, Q9Y2U4 | Gene names | KCNK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7. | |||||
|
KCNK7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.069353 (rank : 56) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z2T1, Q9QXY0, Q9QYE8, Q9R1V1, Q9R242 | Gene names | Kcnk7, Dpkch3, Kcnk6, Kcnk8, Knot1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7 (Putative potassium channel DP3) (Double-pore K(+) channel 3) (Neuromuscular two p domain potassium channel). | |||||
|
KCNKC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.066618 (rank : 57) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HB15 | Gene names | KCNK12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 12 (Tandem pore domain halothane- inhibited potassium channel 2) (THIK-2). | |||||
|
KCNKD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.070904 (rank : 55) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9HB14, Q96E79 | Gene names | KCNK13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNKD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.088200 (rank : 50) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R1P5 | Gene names | Kcnk13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNH3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 148 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
KCNH3_MOUSE
|
||||||
| NC score | 0.994671 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
KCNH8_HUMAN
|
||||||
| NC score | 0.984717 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96L42 | Gene names | KCNH8 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 8 (Voltage-gated potassium channel subunit Kv12.1) (Ether-a-go-go-like potassium channel 3) (ELK channel 3) (ELK3) (ELK1) (hElk1). | |||||
|
KCNH4_HUMAN
|
||||||
| NC score | 0.980859 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
KCNH1_MOUSE
|
||||||
| NC score | 0.961927 (rank : 5) | θ value | 2.35568e-133 (rank : 6) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
KCNH1_HUMAN
|
||||||
| NC score | 0.961732 (rank : 6) | θ value | 5.24793e-133 (rank : 9) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
KCNH5_HUMAN
|
||||||
| NC score | 0.961603 (rank : 7) | θ value | 4.01817e-133 (rank : 8) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
KCNH5_MOUSE
|
||||||
| NC score | 0.961581 (rank : 8) | θ value | 2.35568e-133 (rank : 7) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
KCNH6_HUMAN
|
||||||
| NC score | 0.960920 (rank : 9) | θ value | 2.77624e-142 (rank : 5) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
KCNH2_HUMAN
|
||||||
| NC score | 0.951879 (rank : 10) | θ value | 2.9938e-112 (rank : 11) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
KCNH2_MOUSE
|
||||||
| NC score | 0.951699 (rank : 11) | θ value | 1.02895e-112 (rank : 10) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
KCNH7_MOUSE
|
||||||
| NC score | 0.950001 (rank : 12) | θ value | 2.89973e-107 (rank : 12) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
KCNH7_HUMAN
|
||||||
| NC score | 0.948978 (rank : 13) | θ value | 3.54468e-105 (rank : 13) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
CNGA2_MOUSE
|
||||||
| NC score | 0.806607 (rank : 14) | θ value | 1.28732e-30 (rank : 17) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q62398 | Gene names | Cnga2, Cncg2, Cncg4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated olfactory channel (Cyclic nucleotide-gated cation channel 2) (CNG channel 2) (CNG-2) (CNG2). | |||||
|
CNGA1_HUMAN
|
||||||
| NC score | 0.797659 (rank : 15) | θ value | 1.6813e-30 (rank : 18) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_MOUSE
|
||||||
| NC score | 0.797070 (rank : 16) | θ value | 4.42448e-31 (rank : 16) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JJZ8, Q9WV01 | Gene names | Cnga3, Cng3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA1_MOUSE
|
||||||
| NC score | 0.793801 (rank : 17) | θ value | 1.52067e-31 (rank : 15) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_HUMAN
|
||||||
| NC score | 0.793382 (rank : 18) | θ value | 8.91499e-32 (rank : 14) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16281, Q9UP64 | Gene names | CNGA3, CNCG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
HCN1_HUMAN
|
||||||
| NC score | 0.781330 (rank : 19) | θ value | 2.42779e-29 (rank : 20) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
HCN1_MOUSE
|
||||||
| NC score | 0.777668 (rank : 20) | θ value | 2.42779e-29 (rank : 21) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
HCN3_HUMAN
|
||||||
| NC score | 0.776507 (rank : 21) | θ value | 1.12775e-26 (rank : 26) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
HCN3_MOUSE
|
||||||
| NC score | 0.775874 (rank : 22) | θ value | 6.61148e-27 (rank : 25) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.767830 (rank : 23) | θ value | 5.40856e-29 (rank : 22) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.765714 (rank : 24) | θ value | 1.4233e-29 (rank : 19) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.764949 (rank : 25) | θ value | 5.40856e-29 (rank : 23) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.761466 (rank : 26) | θ value | 7.06379e-29 (rank : 24) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
CNGB3_MOUSE
|
||||||
| NC score | 0.727910 (rank : 27) | θ value | 4.91457e-14 (rank : 29) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JJZ9 | Gene names | Cngb3, Cng6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit) (Cyclic nucleotide-gated channel subunit CNG6). | |||||
|
CNGB3_HUMAN
|
||||||
| NC score | 0.717019 (rank : 28) | θ value | 9.90251e-15 (rank : 28) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CNGB1_HUMAN
|
||||||
| NC score | 0.715553 (rank : 29) | θ value | 3.63628e-17 (rank : 27) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
KCNKH_HUMAN
|
||||||
| NC score | 0.249220 (rank : 30) | θ value | 0.00175202 (rank : 30) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
KCNKF_HUMAN
|
||||||
| NC score | 0.189068 (rank : 31) | θ value | 0.0193708 (rank : 33) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
KCNK3_HUMAN
|
||||||
| NC score | 0.182208 (rank : 32) | θ value | 0.0113563 (rank : 32) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNK2_HUMAN
|
||||||
| NC score | 0.182061 (rank : 33) | θ value | 0.0563607 (rank : 36) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
KCNK2_MOUSE
|
||||||
| NC score | 0.181386 (rank : 34) | θ value | 0.0563607 (rank : 37) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
KCNK3_MOUSE
|
||||||
| NC score | 0.178806 (rank : 35) | θ value | 0.00869519 (rank : 31) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
KCNKA_HUMAN
|
||||||
| NC score | 0.174029 (rank : 36) | θ value | 0.0431538 (rank : 34) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
KCNK6_HUMAN
|
||||||
| NC score | 0.171806 (rank : 37) | θ value | 0.47712 (rank : 55) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
KCNK4_HUMAN
|
||||||
| NC score | 0.166641 (rank : 38) | θ value | 0.21417 (rank : 42) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
KCNKG_HUMAN
|
||||||
| NC score | 0.158686 (rank : 39) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
KCNK9_MOUSE
|
||||||
| NC score | 0.153184 (rank : 40) | θ value | 0.279714 (rank : 47) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q3LS21 | Gene names | Kcnk9, Task3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3). | |||||
|
KCNK9_HUMAN
|
||||||
| NC score | 0.153035 (rank : 41) | θ value | 0.279714 (rank : 46) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NPC2, Q2M290, Q540F2 | Gene names | KCNK9, TASK3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3) (Two pore potassium channel KT3.2). | |||||
|
KCNK4_MOUSE
|
||||||
| NC score | 0.152361 (rank : 42) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
|
KCNK1_HUMAN
|
||||||
| NC score | 0.126119 (rank : 43) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O00180, Q13307 | Gene names | KCNK1, HOHO1, KCNO1, TWIK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1) (Potassium channel KCNO1). | |||||
|
KAP0_HUMAN
|
||||||
| NC score | 0.126066 (rank : 44) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
KCNK5_HUMAN
|
||||||
| NC score | 0.125483 (rank : 45) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O95279 | Gene names | KCNK5, TASK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 5 (Acid-sensitive potassium channel protein TASK-2) (TWIK-related acid-sensitive K(+) channel 2). | |||||
|
KCNK1_MOUSE
|
||||||
| NC score | 0.122328 (rank : 46) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O08581 | Gene names | Kcnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1). | |||||
|
KAP0_MOUSE
|
||||||
| NC score | 0.115004 (rank : 47) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
KAP1_HUMAN
|
||||||
| NC score | 0.104206 (rank : 48) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP1_MOUSE
|
||||||
| NC score | 0.101834 (rank : 49) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KCNKD_MOUSE
|
||||||
| NC score | 0.088200 (rank : 50) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R1P5 | Gene names | Kcnk13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KAP3_MOUSE
|
||||||
| NC score | 0.077118 (rank : 51) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP2_MOUSE
|
||||||
| NC score | 0.075813 (rank : 52) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12367 | Gene names | Prkar2a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP3_HUMAN
|
||||||
| NC score | 0.073902 (rank : 53) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP2_HUMAN
|
||||||
| NC score | 0.073723 (rank : 54) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KCNKD_HUMAN
|
||||||
| NC score | 0.070904 (rank : 55) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9HB14, Q96E79 | Gene names | KCNK13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNK7_MOUSE
|
||||||
| NC score | 0.069353 (rank : 56) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z2T1, Q9QXY0, Q9QYE8, Q9R1V1, Q9R242 | Gene names | Kcnk7, Dpkch3, Kcnk6, Kcnk8, Knot1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7 (Putative potassium channel DP3) (Double-pore K(+) channel 3) (Neuromuscular two p domain potassium channel). | |||||
|
KCNKC_HUMAN
|
||||||
| NC score | 0.066618 (rank : 57) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HB15 | Gene names | KCNK12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 12 (Tandem pore domain halothane- inhibited potassium channel 2) (THIK-2). | |||||
|
CT152_MOUSE
|
||||||
| NC score | 0.059874 (rank : 58) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D5U8, Q8R083 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152 homolog. | |||||
|
KCNK7_HUMAN
|
||||||
| NC score | 0.052615 (rank : 59) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y2U2, Q9Y2U3, Q9Y2U4 | Gene names | KCNK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7. | |||||
|
KCND1_MOUSE
|
||||||
| NC score | 0.046992 (rank : 60) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.043056 (rank : 61) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
KGP1B_HUMAN
|
||||||
| NC score | 0.042853 (rank : 62) | θ value | 0.21417 (rank : 44) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 921 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P14619 | Gene names | PRKG1, PRKG1B, PRKGR1B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
KGP1A_HUMAN
|
||||||
| NC score | 0.042800 (rank : 63) | θ value | 0.21417 (rank : 43) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13976 | Gene names | PRKG1, PRKGR1A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, alpha isozyme (EC 2.7.11.12) (CGK 1 alpha) (cGKI-alpha). | |||||
|
KGP1B_MOUSE
|
||||||
| NC score | 0.042375 (rank : 64) | θ value | 0.47712 (rank : 56) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Z0Z0 | Gene names | Prkg1, Prkg1b, Prkgr1b | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.040390 (rank : 65) | θ value | 0.365318 (rank : 53) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.036709 (rank : 66) | θ value | 0.813845 (rank : 63) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.036260 (rank : 67) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
KCNN4_HUMAN
|
||||||
| NC score | 0.036259 (rank : 68) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15554 | Gene names | KCNN4, IK1, IKCA1, KCA4, SK4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1) (IKCa1) (Putative Gardos channel). | |||||
|
EPN3_HUMAN
|
||||||
| NC score | 0.035038 (rank : 69) | θ value | 0.47712 (rank : 54) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H201, Q9BVN6, Q9NWK2 | Gene names | EPN3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.034905 (rank : 70) | θ value | 0.279714 (rank : 45) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.034663 (rank : 71) | θ value | 0.0961366 (rank : 39) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.034308 (rank : 72) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
KCNN4_MOUSE
|
||||||
| NC score | 0.033355 (rank : 73) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O89109, Q3U1J8, Q9CY39 | Gene names | Kcnn4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1). | |||||
|
TS13_MOUSE
|
||||||
| NC score | 0.032954 (rank : 74) | θ value | 0.47712 (rank : 57) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01755 | Gene names | Tcp11 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific protein PBS13. | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.032166 (rank : 75) | θ value | 0.62314 (rank : 59) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
MINP1_HUMAN
|
||||||
| NC score | 0.032146 (rank : 76) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UNW1, O95172, O95286, Q59EJ2, Q9UGA3 | Gene names | MINPP1, MIPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple inositol polyphosphate phosphatase 1 precursor (EC 3.1.3.62) (Inositol (1,3,4,5)-tetrakisphosphate 3-phosphatase) (Ins(1,3,4,5)P(4) 3-phosphatase). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.031556 (rank : 77) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.030584 (rank : 78) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.028458 (rank : 79) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.027013 (rank : 80) | θ value | 0.365318 (rank : 52) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
HAIR_HUMAN
|
||||||
| NC score | 0.026850 (rank : 81) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
CEP68_HUMAN
|
||||||
| NC score | 0.025124 (rank : 82) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q76N32, O60326, Q9BQ18, Q9UDM9 | Gene names | CEP68, KIAA0582 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.024748 (rank : 83) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
PHLB1_MOUSE
|
||||||
| NC score | 0.023954 (rank : 84) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
CXXC6_HUMAN
|
||||||
| NC score | 0.023436 (rank : 85) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
BIN1_HUMAN
|
||||||
| NC score | 0.023220 (rank : 86) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
CKLF_HUMAN
|
||||||
| NC score | 0.022903 (rank : 87) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBR5, Q9UHM7, Q9UHN8, Q9UI41 | Gene names | CKLF | |||
|
Domain Architecture |

|
|||||
| Description | Chemokine-like factor (C32). | |||||
|
RTP3_MOUSE
|
||||||
| NC score | 0.021772 (rank : 88) | θ value | 0.62314 (rank : 61) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
LCOR_MOUSE
|
||||||
| NC score | 0.021764 (rank : 89) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZPI3, Q3U302, Q5CZW7, Q80VA8, Q8BGT2, Q8C9Q0 | Gene names | Lcor, Kiaa1795, Mlr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ligand-dependent corepressor (LCoR) (Mblk1-related protein 2). | |||||
|
KCNN2_HUMAN
|
||||||
| NC score | 0.021593 (rank : 90) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H2S1 | Gene names | KCNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN2_MOUSE
|
||||||
| NC score | 0.021570 (rank : 91) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P58390 | Gene names | Kcnn2, Sk2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
EPN3_MOUSE
|
||||||
| NC score | 0.020754 (rank : 92) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.020424 (rank : 93) | θ value | 0.279714 (rank : 48) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.020381 (rank : 94) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
BIN1_MOUSE
|
||||||
| NC score | 0.020321 (rank : 95) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08539, Q62434 | Gene names | Bin1, Amphl, Sh3p9 | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (SH3-domain-containing protein 9). | |||||
|
CI079_HUMAN
|
||||||
| NC score | 0.020210 (rank : 96) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
LCOR_HUMAN
|
||||||
| NC score | 0.019948 (rank : 97) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96JN0, Q5VW16, Q7Z723, Q86T32, Q86T33, Q8N3L6 | Gene names | LCOR, KIAA1795, MLR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ligand-dependent corepressor (LCoR) (Mblk1-related protein 2). | |||||
|
CCG8_MOUSE
|
||||||
| NC score | 0.019364 (rank : 98) | θ value | 0.0563607 (rank : 35) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHW2 | Gene names | Cacng8 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-8 subunit (Neuronal voltage- gated calcium channel gamma-8 subunit). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.018868 (rank : 99) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.018638 (rank : 100) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
DAB2_HUMAN
|
||||||
| NC score | 0.018446 (rank : 101) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
PSL1_MOUSE
|
||||||
| NC score | 0.018282 (rank : 102) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3TD49, Q3TLQ8, Q3UG60, Q80VZ6 | Gene names | Sppl2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.018233 (rank : 103) | θ value | 0.365318 (rank : 51) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.017805 (rank : 104) | θ value | 0.365318 (rank : 50) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.017361 (rank : 105) | θ value | 0.163984 (rank : 41) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.017306 (rank : 106) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
FOXN1_HUMAN
|
||||||
| NC score | 0.017278 (rank : 107) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15353, O15352 | Gene names | FOXN1, WHN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude). | |||||
|
PDZD6_HUMAN
|
||||||
| NC score | 0.017252 (rank : 108) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULD6, Q4W5I8, Q86V55 | Gene names | PDZD6, KIAA1284, PDZK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 6. | |||||
|
XKR7_MOUSE
|
||||||
| NC score | 0.017194 (rank : 109) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5GH64 | Gene names | Xkr7, Xrg7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 7. | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.016928 (rank : 110) | θ value | 0.365318 (rank : 49) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CMTA2_MOUSE
|
||||||
| NC score | 0.016866 (rank : 111) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80Y50, Q3TFK0, Q5SX68, Q80TP1, Q8R0D9, Q8R2N5 | Gene names | Camta2, Kiaa0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
CJ108_HUMAN
|
||||||
| NC score | 0.016114 (rank : 112) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N8Z3 | Gene names | C10orf108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative uncharacterized protein C10orf108. | |||||
|
BICD1_HUMAN
|
||||||
| NC score | 0.015975 (rank : 113) | θ value | 0.125558 (rank : 40) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
IRS2_MOUSE
|
||||||
| NC score | 0.015859 (rank : 114) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.015517 (rank : 115) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.015473 (rank : 116) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
ZN703_HUMAN
|
||||||
| NC score | 0.014751 (rank : 117) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H7S9, Q5XG76 | Gene names | ZNF703 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 703. | |||||
|
SENP3_MOUSE
|
||||||
| NC score | 0.014601 (rank : 118) | θ value | 0.813845 (rank : 64) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EP97 | Gene names | Senp3, Smt3ip, Smt3ip1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 3 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP3) (SUMO-1-specific protease 3) (Smt3-specific isopeptidase 1) (Smt3ip1). | |||||
|
EPN2_MOUSE
|
||||||
| NC score | 0.014473 (rank : 119) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
|
JPH2_MOUSE
|
||||||
| NC score | 0.013811 (rank : 120) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
FOXN1_MOUSE
|
||||||
| NC score | 0.013680 (rank : 121) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
BICD1_MOUSE
|
||||||
| NC score | 0.012589 (rank : 122) | θ value | 0.62314 (rank : 58) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
JPH1_MOUSE
|
||||||
| NC score | 0.012519 (rank : 123) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ET80, Q9EQZ3 | Gene names | Jph1, Jp1 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
GA2L2_MOUSE
|
||||||
| NC score | 0.012166 (rank : 124) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.011729 (rank : 125) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
CK056_HUMAN
|
||||||
| NC score | 0.011023 (rank : 126) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N612, Q9C0A4, Q9H0N3, Q9H624 | Gene names | C11orf56, KIAA1759 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf56. | |||||
|
K0179_HUMAN
|
||||||
| NC score | 0.010943 (rank : 127) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14684, Q8TBZ4 | Gene names | KIAA0179 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0179. | |||||
|
PK2L1_HUMAN
|
||||||
| NC score | 0.010698 (rank : 128) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
APXL_HUMAN
|
||||||
| NC score | 0.010323 (rank : 129) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
LIMD1_MOUSE
|
||||||
| NC score | 0.010196 (rank : 130) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.010160 (rank : 131) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
HD_HUMAN
|
||||||
| NC score | 0.009071 (rank : 132) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
NBEA_MOUSE
|
||||||
| NC score | 0.008931 (rank : 133) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EPN1, Q8C931, Q9EPM9, Q9EPN0, Q9WVM9 | Gene names | Nbea, Lyst2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2). | |||||
|
NBEA_HUMAN
|
||||||
| NC score | 0.008818 (rank : 134) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NFP9, Q9HCM8, Q9NSU1, Q9NW98, Q9Y6J1 | Gene names | NBEA, BCL8B, KIAA1544, LYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2) (Protein BCL8B). | |||||
|
SEM6B_HUMAN
|
||||||
| NC score | 0.008367 (rank : 135) | θ value | 0.62314 (rank : 62) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H3T3, Q9NRK9 | Gene names | SEMA6B, SEMAZ | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin Z) (Sema Z). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.007916 (rank : 136) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.007406 (rank : 137) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
WD21C_HUMAN
|
||||||
| NC score | 0.007335 (rank : 138) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NA75 | Gene names | WDR21C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 21C. | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.007058 (rank : 139) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
PLXB1_MOUSE
|
||||||
| NC score | 0.006715 (rank : 140) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.006517 (rank : 141) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
INVS_MOUSE
|
||||||
| NC score | 0.006408 (rank : 142) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
SUSD4_MOUSE
|
||||||
| NC score | 0.006143 (rank : 143) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BH32, Q8VC43 | Gene names | Susd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
DUS2_HUMAN
|
||||||
| NC score | 0.005672 (rank : 144) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q05923 | Gene names | DUSP2, PAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
EMID1_MOUSE
|
||||||
| NC score | 0.005194 (rank : 145) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91VF5 | Gene names | Emid1, Emu1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EMI domain-containing protein 1 precursor (Protein Emu1) (Emilin and multimerin domain-containing protein 1). | |||||
|
MAST2_HUMAN
|
||||||
| NC score | 0.004396 (rank : 146) | θ value | 0.0736092 (rank : 38) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
EPAS1_MOUSE
|
||||||
| NC score | 0.004394 (rank : 147) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
PANK1_MOUSE
|
||||||
| NC score | 0.003904 (rank : 148) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K4K6, Q9D3K1, Q9QXM8 | Gene names | Pank1, Pank | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (mPank1) (mPank). | |||||
|
PTC2_HUMAN
|
||||||
| NC score | 0.003762 (rank : 149) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6C5, O95341, O95856, Q5QP87, Q6UX14 | Gene names | PTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 2 (PTC2). | |||||
|
CAMKV_HUMAN
|
||||||
| NC score | 0.003255 (rank : 150) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.001724 (rank : 151) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
DGKG_MOUSE
|
||||||
| NC score | 0.001362 (rank : 152) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91WG7 | Gene names | Dgkg, Dagk3 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase gamma (EC 2.7.1.107) (Diglyceride kinase gamma) (DGK-gamma) (DAG kinase gamma) (88 kDa diacylglycerol kinase). | |||||
|
KKCC1_HUMAN
|
||||||
| NC score | 0.000567 (rank : 153) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N5S9, Q9BQH3 | Gene names | CAMKK1, CAMKKA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase kinase 1 (EC 2.7.11.17) (Calcium/calmodulin-dependent protein kinase kinase alpha) (CaM-kinase kinase alpha) (CaM-KK alpha) (CaMKK alpha) (CaMKK 1) (CaM-kinase IV kinase). | |||||
|
KSR1_MOUSE
|
||||||
| NC score | -0.000022 (rank : 154) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61097, Q61648, Q78DX8 | Gene names | Ksr1, Ksr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras) (mKSR1) (Hb protein). | |||||
|
STK10_HUMAN
|
||||||
| NC score | -0.000650 (rank : 155) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
M3K10_HUMAN
|
||||||
| NC score | -0.000873 (rank : 156) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q02779, Q12761, Q14871 | Gene names | MAP3K10, MLK2, MST | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 10 (EC 2.7.11.25) (Mixed lineage kinase 2) (Protein kinase MST). | |||||
|
VASN_HUMAN
|
||||||
| NC score | -0.001514 (rank : 157) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6EMK4, Q6UXL4, Q6UXL5, Q96CX1 | Gene names | VASN, SLITL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vasorin precursor (Protein Slit-like 2). | |||||
|
EGR4_HUMAN
|
||||||
| NC score | -0.002502 (rank : 158) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
MTR1L_HUMAN
|
||||||
| NC score | -0.002633 (rank : 159) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1180 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13585 | Gene names | GPR50 | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin-related receptor (G protein-coupled receptor 50) (H9). | |||||
|
ZF276_HUMAN
|
||||||
| NC score | -0.002805 (rank : 160) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N554 | Gene names | ZFP276 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 276 homolog (Zfp-276). | |||||
|
ZN227_HUMAN
|
||||||
| NC score | -0.003956 (rank : 161) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 781 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86WZ6 | Gene names | ZNF227 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 227. | |||||
|
ZN169_HUMAN
|
||||||
| NC score | -0.004091 (rank : 162) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14929, Q6PI28 | Gene names | ZNF169 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 169. | |||||