Please be patient as the page loads
|
KCNH3_MOUSE
|
||||||
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
KCNH3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994671 (rank : 2) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
KCNH3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 133 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
KCNH4_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.980755 (rank : 4) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
KCNH8_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.984758 (rank : 3) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96L42 | Gene names | KCNH8 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 8 (Voltage-gated potassium channel subunit Kv12.1) (Ether-a-go-go-like potassium channel 3) (ELK channel 3) (ELK3) (ELK1) (hElk1). | |||||
|
KCNH6_HUMAN
|
||||||
| θ value | 5.23575e-141 (rank : 5) | NC score | 0.961308 (rank : 9) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
KCNH1_MOUSE
|
||||||
| θ value | 4.01817e-133 (rank : 6) | NC score | 0.961886 (rank : 5) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
KCNH5_MOUSE
|
||||||
| θ value | 5.24793e-133 (rank : 7) | NC score | 0.961543 (rank : 7) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
KCNH1_HUMAN
|
||||||
| θ value | 8.9516e-133 (rank : 8) | NC score | 0.961757 (rank : 6) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
KCNH5_HUMAN
|
||||||
| θ value | 8.9516e-133 (rank : 9) | NC score | 0.961425 (rank : 8) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
KCNH2_MOUSE
|
||||||
| θ value | 1.75513e-112 (rank : 10) | NC score | 0.951348 (rank : 11) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
KCNH2_HUMAN
|
||||||
| θ value | 2.29227e-112 (rank : 11) | NC score | 0.951447 (rank : 10) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
KCNH7_MOUSE
|
||||||
| θ value | 2.22025e-107 (rank : 12) | NC score | 0.950083 (rank : 12) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
KCNH7_HUMAN
|
||||||
| θ value | 2.71407e-105 (rank : 13) | NC score | 0.948987 (rank : 13) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
CNGA1_MOUSE
|
||||||
| θ value | 1.37539e-32 (rank : 14) | NC score | 0.793841 (rank : 17) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_MOUSE
|
||||||
| θ value | 6.82597e-32 (rank : 15) | NC score | 0.797120 (rank : 16) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JJZ8, Q9WV01 | Gene names | Cnga3, Cng3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA2_MOUSE
|
||||||
| θ value | 8.91499e-32 (rank : 16) | NC score | 0.806599 (rank : 14) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q62398 | Gene names | Cnga2, Cncg2, Cncg4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated olfactory channel (Cyclic nucleotide-gated cation channel 2) (CNG channel 2) (CNG-2) (CNG2). | |||||
|
CNGA1_HUMAN
|
||||||
| θ value | 1.16434e-31 (rank : 17) | NC score | 0.797803 (rank : 15) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 8.3442e-30 (rank : 18) | NC score | 0.764495 (rank : 24) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
HCN1_HUMAN
|
||||||
| θ value | 2.42779e-29 (rank : 19) | NC score | 0.781208 (rank : 19) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
HCN1_MOUSE
|
||||||
| θ value | 2.42779e-29 (rank : 20) | NC score | 0.777258 (rank : 20) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
CNGA3_HUMAN
|
||||||
| θ value | 3.17079e-29 (rank : 21) | NC score | 0.793261 (rank : 18) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16281, Q9UP64 | Gene names | CNGA3, CNCG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 4.14116e-29 (rank : 22) | NC score | 0.759634 (rank : 26) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 5.40856e-29 (rank : 23) | NC score | 0.762826 (rank : 25) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
HCN2_MOUSE
|
||||||
| θ value | 2.68423e-28 (rank : 24) | NC score | 0.766645 (rank : 23) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
HCN3_MOUSE
|
||||||
| θ value | 1.47289e-26 (rank : 25) | NC score | 0.775849 (rank : 21) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
HCN3_HUMAN
|
||||||
| θ value | 3.28125e-26 (rank : 26) | NC score | 0.775828 (rank : 22) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
CNGB1_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 27) | NC score | 0.714034 (rank : 29) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CNGB3_HUMAN
|
||||||
| θ value | 4.91457e-14 (rank : 28) | NC score | 0.715687 (rank : 28) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CNGB3_MOUSE
|
||||||
| θ value | 5.43371e-13 (rank : 29) | NC score | 0.726821 (rank : 27) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JJZ9 | Gene names | Cngb3, Cng6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit) (Cyclic nucleotide-gated channel subunit CNG6). | |||||
|
KCNKH_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 30) | NC score | 0.250350 (rank : 30) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 31) | NC score | 0.038091 (rank : 67) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
KCNK3_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 32) | NC score | 0.179610 (rank : 35) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
KCNK3_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 33) | NC score | 0.182977 (rank : 33) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNKF_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 34) | NC score | 0.189924 (rank : 31) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 35) | NC score | 0.051520 (rank : 60) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 36) | NC score | 0.038772 (rank : 66) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 37) | NC score | 0.035458 (rank : 70) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
KCNK4_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 38) | NC score | 0.168936 (rank : 38) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
KCNKA_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 39) | NC score | 0.175337 (rank : 36) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
MINP1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 40) | NC score | 0.049635 (rank : 61) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UNW1, O95172, O95286, Q59EJ2, Q9UGA3 | Gene names | MINPP1, MIPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple inositol polyphosphate phosphatase 1 precursor (EC 3.1.3.62) (Inositol (1,3,4,5)-tetrakisphosphate 3-phosphatase) (Ins(1,3,4,5)P(4) 3-phosphatase). | |||||
|
KCNK2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 41) | NC score | 0.183384 (rank : 32) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
KCNK2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 42) | NC score | 0.182717 (rank : 34) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
KGP1A_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 43) | NC score | 0.041923 (rank : 64) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13976 | Gene names | PRKG1, PRKGR1A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, alpha isozyme (EC 2.7.11.12) (CGK 1 alpha) (cGKI-alpha). | |||||
|
KGP1B_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 44) | NC score | 0.041989 (rank : 63) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 921 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P14619 | Gene names | PRKG1, PRKG1B, PRKGR1B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
MTA2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 45) | NC score | 0.021233 (rank : 88) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R190, Q3TVT8 | Gene names | Mta2, Mta1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1). | |||||
|
BICD1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 46) | NC score | 0.015786 (rank : 102) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
KGP1B_MOUSE
|
||||||
| θ value | 0.163984 (rank : 47) | NC score | 0.041510 (rank : 65) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z0Z0 | Gene names | Prkg1, Prkg1b, Prkgr1b | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
KCNK9_HUMAN
|
||||||
| θ value | 0.279714 (rank : 48) | NC score | 0.153868 (rank : 42) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NPC2, Q2M290, Q540F2 | Gene names | KCNK9, TASK3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3) (Two pore potassium channel KT3.2). | |||||
|
KCNK9_MOUSE
|
||||||
| θ value | 0.279714 (rank : 49) | NC score | 0.154050 (rank : 40) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q3LS21 | Gene names | Kcnk9, Task3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3). | |||||
|
FOXN1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | 0.013901 (rank : 106) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
KCNK6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 51) | NC score | 0.172856 (rank : 37) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.021544 (rank : 87) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
KCNK5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.126564 (rank : 45) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O95279 | Gene names | KCNK5, TASK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 5 (Acid-sensitive potassium channel protein TASK-2) (TWIK-related acid-sensitive K(+) channel 2). | |||||
|
SEM6C_MOUSE
|
||||||
| θ value | 0.62314 (rank : 54) | NC score | 0.011659 (rank : 114) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
BICD1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.012342 (rank : 110) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
DCBD1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.012200 (rank : 111) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D4J3, Q8R327, Q9D696 | Gene names | Dcbld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
DEPP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.025240 (rank : 81) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2F3, Q8BU66 | Gene names | Depp, Fseg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEPP (Fat-specific expressed gene protein). | |||||
|
FBX46_MOUSE
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.017653 (rank : 94) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BG80 | Gene names | Fbxo46, Fbx46, Fbxo34l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
KCNK4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.154005 (rank : 41) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
|
KCNKG_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.159805 (rank : 39) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.009776 (rank : 120) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
RFTN1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.015830 (rank : 101) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14699, Q496Y2, Q4QQI7, Q5JB48, Q7Z7P2 | Gene names | RFTN1, KIAA0084 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Raftlin (Raft-linking protein) (Cell migration-inducing gene 2 protein). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.017056 (rank : 98) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.007320 (rank : 126) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.027471 (rank : 77) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
MTA2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.017208 (rank : 97) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94776, Q9UQB5 | Gene names | MTA2, MTA1L1, PID | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1) (MTA1-L1 protein) (p53 target protein in deacetylase complex). | |||||
|
CASZ1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.012811 (rank : 108) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 473 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86V15, Q5T9S1, Q8WX49, Q8WX50, Q9BT16, Q9NXC6 | Gene names | CASZ1, CST, ZNF693 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Castor homolog 1 zinc finger protein (Castor-related protein) (Zinc finger protein 693). | |||||
|
COIA1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.010434 (rank : 116) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
GA2L2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.014554 (rank : 105) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
IRS2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.018596 (rank : 91) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
MINP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.031546 (rank : 73) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2L6, Q8VDR0, Q9JJD5 | Gene names | Minpp1, Mipp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple inositol polyphosphate phosphatase 1 precursor (EC 3.1.3.62) (Inositol (1,3,4,5)-tetrakisphosphate 3-phosphatase) (Ins(1,3,4,5)P(4) 3-phosphatase). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.024950 (rank : 82) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
PHLB1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.025998 (rank : 80) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
EPN3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.028792 (rank : 75) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H201, Q9BVN6, Q9NWK2 | Gene names | EPN3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
HAIR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.026843 (rank : 78) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
KCNN4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.036333 (rank : 68) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15554 | Gene names | KCNN4, IK1, IKCA1, KCA4, SK4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1) (IKCa1) (Putative Gardos channel). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.024473 (rank : 83) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.036038 (rank : 69) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.006603 (rank : 130) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
SI1L3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.010683 (rank : 115) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
ATS13_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.002560 (rank : 139) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
DAB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.017341 (rank : 95) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
KCNK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.127096 (rank : 44) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O00180, Q13307 | Gene names | KCNK1, HOHO1, KCNO1, TWIK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1) (Potassium channel KCNO1). | |||||
|
KCNN4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.033418 (rank : 71) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O89109, Q3U1J8, Q9CY39 | Gene names | Kcnn4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1). | |||||
|
PSL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.026686 (rank : 79) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3TD49, Q3TLQ8, Q3UG60, Q80VZ6 | Gene names | Sppl2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b). | |||||
|
TSC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.006245 (rank : 131) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92574 | Gene names | TSC1, KIAA0243, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Hamartin (Tuberous sclerosis 1 protein). | |||||
|
XKR7_MOUSE
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.017309 (rank : 96) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5GH64 | Gene names | Xkr7, Xrg7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 7. | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.002808 (rank : 138) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.033105 (rank : 72) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CAMKV_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.001534 (rank : 142) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
GRIN1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.016352 (rank : 100) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.010043 (rank : 119) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
KCNK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.123411 (rank : 46) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O08581 | Gene names | Kcnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.012111 (rank : 112) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.005123 (rank : 133) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | -0.000241 (rank : 145) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
PK2L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.010384 (rank : 117) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.028230 (rank : 76) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
ADM2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.015616 (rank : 103) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TNK8 | Gene names | Adm2, Am2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADM2 precursor (Intermedin) [Contains: Adrenomedullin-2 (Intermedin- long) (IMDL); Intermedin-short (IMDS)]. | |||||
|
DUS2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.005703 (rank : 132) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q05923 | Gene names | DUSP2, PAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
K0423_MOUSE
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.007873 (rank : 123) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6A070 | Gene names | Kiaa0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
KIRR2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.001501 (rank : 143) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TSU7, Q7TQ98 | Gene names | Kirrel2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kin of IRRE-like protein 2 precursor (Kin of irregular chiasm-like protein 2). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.000370 (rank : 144) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.019258 (rank : 89) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PHLB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.019184 (rank : 90) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86UU1, O75133, Q4KMF8, Q8TEQ2 | Gene names | PHLDB1, DLNB07, KIAA0638, LL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
RBNS5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.006702 (rank : 128) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80Y56, Q8K0L6, Q9CTW0 | Gene names | Zfyve20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20). | |||||
|
RUNX1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.007645 (rank : 124) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q03347, O08598, Q62049, Q9ESB9, Q9ET65 | Gene names | Runx1, Aml1, Cbfa2, Pebp2ab | |||
|
Domain Architecture |
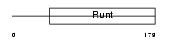
|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
TNR4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.006700 (rank : 129) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P43489, Q13663, Q5T7M0 | Gene names | TNFRSF4, TXGP1L | |||
|
Domain Architecture |
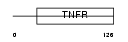
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 4 precursor (OX40L receptor) (ACT35 antigen) (TAX transcriptionally-activated glycoprotein 1 receptor) (CD134 antigen). | |||||
|
ARHG3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.002926 (rank : 137) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NR81, Q6NUN3, Q7Z4U2, Q7Z5T2, Q9H7T4 | Gene names | ARHGEF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 3 (Exchange factor found in platelets and leukemic and neuronal tissues) (XPLN). | |||||
|
BIN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.016711 (rank : 99) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
BIN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.015052 (rank : 104) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08539, Q62434 | Gene names | Bin1, Amphl, Sh3p9 | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (SH3-domain-containing protein 9). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.006896 (rank : 127) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.004752 (rank : 135) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CKLF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.023639 (rank : 84) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBR5, Q9UHM7, Q9UHN8, Q9UI41 | Gene names | CKLF | |||
|
Domain Architecture |

|
|||||
| Description | Chemokine-like factor (C32). | |||||
|
CMTA2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.012360 (rank : 109) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80Y50, Q3TFK0, Q5SX68, Q80TP1, Q8R0D9, Q8R2N5 | Gene names | Camta2, Kiaa0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
HD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.010248 (rank : 118) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
KCND1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.046861 (rank : 62) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCNN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.021575 (rank : 85) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H2S1 | Gene names | KCNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.021552 (rank : 86) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P58390 | Gene names | Kcnn2, Sk2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.009683 (rank : 121) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SLY_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.004987 (rank : 134) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K352, Q3TBX0, Q9DBV2 | Gene names | Sly | |||
|
Domain Architecture |
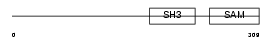
|
|||||
| Description | SH3 protein expressed in lymphocytes. | |||||
|
TBX2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.004541 (rank : 136) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13207, Q16424, Q7Z647 | Gene names | TBX2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
TENS4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.013679 (rank : 107) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IZW8, Q71RB7, Q8WV64, Q96JV4 | Gene names | TNS4, CTEN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor (C-terminal tensin-like protein). | |||||
|
ZN227_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | -0.003956 (rank : 146) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 781 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86WZ6 | Gene names | ZNF227 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 227. | |||||
|
CXXC6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.018131 (rank : 93) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
FOXN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.018427 (rank : 92) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15353, O15352 | Gene names | FOXN1, WHN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude). | |||||
|
HCLS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.001873 (rank : 140) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49710 | Gene names | Hcls1, Hs1 | |||
|
Domain Architecture |

|
|||||
| Description | Hematopoietic lineage cell-specific protein (Hematopoietic cell- specific LYN substrate 1) (LckBP1). | |||||
|
LIMD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.008561 (rank : 122) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.011985 (rank : 113) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.029966 (rank : 74) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
WD21C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.007372 (rank : 125) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NA75 | Gene names | WDR21C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 21C. | |||||
|
ZN169_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | -0.004128 (rank : 147) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14929, Q6PI28 | Gene names | ZNF169 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 169. | |||||
|
ZO2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.001760 (rank : 141) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0U1, Q8K210 | Gene names | Tjp2, Zo2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
CT152_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.061134 (rank : 58) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D5U8, Q8R083 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152 homolog. | |||||
|
KAP0_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.127812 (rank : 43) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
KAP0_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.116809 (rank : 47) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
KAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.106012 (rank : 48) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.103640 (rank : 49) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.075471 (rank : 54) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.077593 (rank : 52) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12367 | Gene names | Prkar2a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.075677 (rank : 53) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.078890 (rank : 51) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KCNK7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.053452 (rank : 59) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y2U2, Q9Y2U3, Q9Y2U4 | Gene names | KCNK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7. | |||||
|
KCNK7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.070182 (rank : 56) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z2T1, Q9QXY0, Q9QYE8, Q9R1V1, Q9R242 | Gene names | Kcnk7, Dpkch3, Kcnk6, Kcnk8, Knot1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7 (Putative potassium channel DP3) (Double-pore K(+) channel 3) (Neuromuscular two p domain potassium channel). | |||||
|
KCNKC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.067371 (rank : 57) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HB15 | Gene names | KCNK12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 12 (Tandem pore domain halothane- inhibited potassium channel 2) (THIK-2). | |||||
|
KCNKD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.071647 (rank : 55) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9HB14, Q96E79 | Gene names | KCNK13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNKD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.088946 (rank : 50) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R1P5 | Gene names | Kcnk13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNH3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 133 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
KCNH3_HUMAN
|
||||||
| NC score | 0.994671 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
KCNH8_HUMAN
|
||||||
| NC score | 0.984758 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96L42 | Gene names | KCNH8 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 8 (Voltage-gated potassium channel subunit Kv12.1) (Ether-a-go-go-like potassium channel 3) (ELK channel 3) (ELK3) (ELK1) (hElk1). | |||||
|
KCNH4_HUMAN
|
||||||
| NC score | 0.980755 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
KCNH1_MOUSE
|
||||||
| NC score | 0.961886 (rank : 5) | θ value | 4.01817e-133 (rank : 6) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
KCNH1_HUMAN
|
||||||
| NC score | 0.961757 (rank : 6) | θ value | 8.9516e-133 (rank : 8) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
KCNH5_MOUSE
|
||||||
| NC score | 0.961543 (rank : 7) | θ value | 5.24793e-133 (rank : 7) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
KCNH5_HUMAN
|
||||||
| NC score | 0.961425 (rank : 8) | θ value | 8.9516e-133 (rank : 9) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
KCNH6_HUMAN
|
||||||
| NC score | 0.961308 (rank : 9) | θ value | 5.23575e-141 (rank : 5) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
KCNH2_HUMAN
|
||||||
| NC score | 0.951447 (rank : 10) | θ value | 2.29227e-112 (rank : 11) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
KCNH2_MOUSE
|
||||||
| NC score | 0.951348 (rank : 11) | θ value | 1.75513e-112 (rank : 10) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
KCNH7_MOUSE
|
||||||
| NC score | 0.950083 (rank : 12) | θ value | 2.22025e-107 (rank : 12) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
KCNH7_HUMAN
|
||||||
| NC score | 0.948987 (rank : 13) | θ value | 2.71407e-105 (rank : 13) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
CNGA2_MOUSE
|
||||||
| NC score | 0.806599 (rank : 14) | θ value | 8.91499e-32 (rank : 16) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q62398 | Gene names | Cnga2, Cncg2, Cncg4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated olfactory channel (Cyclic nucleotide-gated cation channel 2) (CNG channel 2) (CNG-2) (CNG2). | |||||
|
CNGA1_HUMAN
|
||||||
| NC score | 0.797803 (rank : 15) | θ value | 1.16434e-31 (rank : 17) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_MOUSE
|
||||||
| NC score | 0.797120 (rank : 16) | θ value | 6.82597e-32 (rank : 15) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JJZ8, Q9WV01 | Gene names | Cnga3, Cng3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA1_MOUSE
|
||||||
| NC score | 0.793841 (rank : 17) | θ value | 1.37539e-32 (rank : 14) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_HUMAN
|
||||||
| NC score | 0.793261 (rank : 18) | θ value | 3.17079e-29 (rank : 21) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16281, Q9UP64 | Gene names | CNGA3, CNCG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
HCN1_HUMAN
|
||||||
| NC score | 0.781208 (rank : 19) | θ value | 2.42779e-29 (rank : 19) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
HCN1_MOUSE
|
||||||
| NC score | 0.777258 (rank : 20) | θ value | 2.42779e-29 (rank : 20) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
HCN3_MOUSE
|
||||||
| NC score | 0.775849 (rank : 21) | θ value | 1.47289e-26 (rank : 25) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
HCN3_HUMAN
|
||||||
| NC score | 0.775828 (rank : 22) | θ value | 3.28125e-26 (rank : 26) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.766645 (rank : 23) | θ value | 2.68423e-28 (rank : 24) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.764495 (rank : 24) | θ value | 8.3442e-30 (rank : 18) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.762826 (rank : 25) | θ value | 5.40856e-29 (rank : 23) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.759634 (rank : 26) | θ value | 4.14116e-29 (rank : 22) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
CNGB3_MOUSE
|
||||||
| NC score | 0.726821 (rank : 27) | θ value | 5.43371e-13 (rank : 29) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JJZ9 | Gene names | Cngb3, Cng6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit) (Cyclic nucleotide-gated channel subunit CNG6). | |||||
|
CNGB3_HUMAN
|
||||||
| NC score | 0.715687 (rank : 28) | θ value | 4.91457e-14 (rank : 28) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CNGB1_HUMAN
|
||||||
| NC score | 0.714034 (rank : 29) | θ value | 1.63225e-17 (rank : 27) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
KCNKH_HUMAN
|
||||||
| NC score | 0.250350 (rank : 30) | θ value | 0.00175202 (rank : 30) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
KCNKF_HUMAN
|
||||||
| NC score | 0.189924 (rank : 31) | θ value | 0.0193708 (rank : 34) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
KCNK2_HUMAN
|
||||||
| NC score | 0.183384 (rank : 32) | θ value | 0.0563607 (rank : 41) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
KCNK3_HUMAN
|
||||||
| NC score | 0.182977 (rank : 33) | θ value | 0.0113563 (rank : 33) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNK2_MOUSE
|
||||||
| NC score | 0.182717 (rank : 34) | θ value | 0.0563607 (rank : 42) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
KCNK3_MOUSE
|
||||||
| NC score | 0.179610 (rank : 35) | θ value | 0.00869519 (rank : 32) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
KCNKA_HUMAN
|
||||||
| NC score | 0.175337 (rank : 36) | θ value | 0.0431538 (rank : 39) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
KCNK6_HUMAN
|
||||||
| NC score | 0.172856 (rank : 37) | θ value | 0.47712 (rank : 51) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
KCNK4_HUMAN
|
||||||
| NC score | 0.168936 (rank : 38) | θ value | 0.0431538 (rank : 38) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
KCNKG_HUMAN
|
||||||
| NC score | 0.159805 (rank : 39) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
KCNK9_MOUSE
|
||||||
| NC score | 0.154050 (rank : 40) | θ value | 0.279714 (rank : 49) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q3LS21 | Gene names | Kcnk9, Task3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3). | |||||
|
KCNK4_MOUSE
|
||||||
| NC score | 0.154005 (rank : 41) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
|
KCNK9_HUMAN
|
||||||
| NC score | 0.153868 (rank : 42) | θ value | 0.279714 (rank : 48) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NPC2, Q2M290, Q540F2 | Gene names | KCNK9, TASK3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3) (Two pore potassium channel KT3.2). | |||||
|
KAP0_HUMAN
|
||||||
| NC score | 0.127812 (rank : 43) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
KCNK1_HUMAN
|
||||||
| NC score | 0.127096 (rank : 44) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O00180, Q13307 | Gene names | KCNK1, HOHO1, KCNO1, TWIK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1) (Potassium channel KCNO1). | |||||
|
KCNK5_HUMAN
|
||||||
| NC score | 0.126564 (rank : 45) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O95279 | Gene names | KCNK5, TASK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 5 (Acid-sensitive potassium channel protein TASK-2) (TWIK-related acid-sensitive K(+) channel 2). | |||||
|
KCNK1_MOUSE
|
||||||
| NC score | 0.123411 (rank : 46) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O08581 | Gene names | Kcnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1). | |||||
|
KAP0_MOUSE
|
||||||
| NC score | 0.116809 (rank : 47) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
KAP1_HUMAN
|
||||||
| NC score | 0.106012 (rank : 48) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP1_MOUSE
|
||||||
| NC score | 0.103640 (rank : 49) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KCNKD_MOUSE
|
||||||
| NC score | 0.088946 (rank : 50) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R1P5 | Gene names | Kcnk13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KAP3_MOUSE
|
||||||
| NC score | 0.078890 (rank : 51) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP2_MOUSE
|
||||||
| NC score | 0.077593 (rank : 52) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12367 | Gene names | Prkar2a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP3_HUMAN
|
||||||
| NC score | 0.075677 (rank : 53) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP2_HUMAN
|
||||||
| NC score | 0.075471 (rank : 54) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KCNKD_HUMAN
|
||||||
| NC score | 0.071647 (rank : 55) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9HB14, Q96E79 | Gene names | KCNK13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNK7_MOUSE
|
||||||
| NC score | 0.070182 (rank : 56) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z2T1, Q9QXY0, Q9QYE8, Q9R1V1, Q9R242 | Gene names | Kcnk7, Dpkch3, Kcnk6, Kcnk8, Knot1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7 (Putative potassium channel DP3) (Double-pore K(+) channel 3) (Neuromuscular two p domain potassium channel). | |||||
|
KCNKC_HUMAN
|
||||||
| NC score | 0.067371 (rank : 57) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HB15 | Gene names | KCNK12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 12 (Tandem pore domain halothane- inhibited potassium channel 2) (THIK-2). | |||||
|
CT152_MOUSE
|
||||||
| NC score | 0.061134 (rank : 58) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D5U8, Q8R083 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152 homolog. | |||||
|
KCNK7_HUMAN
|
||||||
| NC score | 0.053452 (rank : 59) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y2U2, Q9Y2U3, Q9Y2U4 | Gene names | KCNK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7. | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.051520 (rank : 60) | θ value | 0.0252991 (rank : 35) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
MINP1_HUMAN
|
||||||
| NC score | 0.049635 (rank : 61) | θ value | 0.0431538 (rank : 40) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UNW1, O95172, O95286, Q59EJ2, Q9UGA3 | Gene names | MINPP1, MIPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple inositol polyphosphate phosphatase 1 precursor (EC 3.1.3.62) (Inositol (1,3,4,5)-tetrakisphosphate 3-phosphatase) (Ins(1,3,4,5)P(4) 3-phosphatase). | |||||
|
KCND1_MOUSE
|
||||||
| NC score | 0.046861 (rank : 62) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KGP1B_HUMAN
|
||||||
| NC score | 0.041989 (rank : 63) | θ value | 0.0736092 (rank : 44) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 921 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P14619 | Gene names | PRKG1, PRKG1B, PRKGR1B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
KGP1A_HUMAN
|
||||||
| NC score | 0.041923 (rank : 64) | θ value | 0.0736092 (rank : 43) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13976 | Gene names | PRKG1, PRKGR1A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, alpha isozyme (EC 2.7.11.12) (CGK 1 alpha) (cGKI-alpha). | |||||
|
KGP1B_MOUSE
|
||||||
| NC score | 0.041510 (rank : 65) | θ value | 0.163984 (rank : 47) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z0Z0 | Gene names | Prkg1, Prkg1b, Prkgr1b | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.038772 (rank : 66) | θ value | 0.0252991 (rank : 36) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.038091 (rank : 67) | θ value | 0.00298849 (rank : 31) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
KCNN4_HUMAN
|
||||||
| NC score | 0.036333 (rank : 68) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15554 | Gene names | KCNN4, IK1, IKCA1, KCA4, SK4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1) (IKCa1) (Putative Gardos channel). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.036038 (rank : 69) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.035458 (rank : 70) | θ value | 0.0330416 (rank : 37) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
KCNN4_MOUSE
|
||||||
| NC score | 0.033418 (rank : 71) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O89109, Q3U1J8, Q9CY39 | Gene names | Kcnn4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.033105 (rank : 72) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
MINP1_MOUSE
|
||||||
| NC score | 0.031546 (rank : 73) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2L6, Q8VDR0, Q9JJD5 | Gene names | Minpp1, Mipp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple inositol polyphosphate phosphatase 1 precursor (EC 3.1.3.62) (Inositol (1,3,4,5)-tetrakisphosphate 3-phosphatase) (Ins(1,3,4,5)P(4) 3-phosphatase). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.029966 (rank : 74) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
EPN3_HUMAN
|
||||||
| NC score | 0.028792 (rank : 75) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H201, Q9BVN6, Q9NWK2 | Gene names | EPN3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.028230 (rank : 76) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.027471 (rank : 77) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
HAIR_HUMAN
|
||||||
| NC score | 0.026843 (rank : 78) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
PSL1_MOUSE
|
||||||
| NC score | 0.026686 (rank : 79) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3TD49, Q3TLQ8, Q3UG60, Q80VZ6 | Gene names | Sppl2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b). | |||||
|
PHLB1_MOUSE
|
||||||
| NC score | 0.025998 (rank : 80) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
DEPP_MOUSE
|
||||||
| NC score | 0.025240 (rank : 81) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2F3, Q8BU66 | Gene names | Depp, Fseg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEPP (Fat-specific expressed gene protein). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.024950 (rank : 82) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.024473 (rank : 83) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
CKLF_HUMAN
|
||||||
| NC score | 0.023639 (rank : 84) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBR5, Q9UHM7, Q9UHN8, Q9UI41 | Gene names | CKLF | |||
|
Domain Architecture |

|
|||||
| Description | Chemokine-like factor (C32). | |||||
|
KCNN2_HUMAN
|
||||||
| NC score | 0.021575 (rank : 85) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H2S1 | Gene names | KCNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN2_MOUSE
|
||||||
| NC score | 0.021552 (rank : 86) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P58390 | Gene names | Kcnn2, Sk2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.021544 (rank : 87) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
MTA2_MOUSE
|
||||||
| NC score | 0.021233 (rank : 88) | θ value | 0.0961366 (rank : 45) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R190, Q3TVT8 | Gene names | Mta2, Mta1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.019258 (rank : 89) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PHLB1_HUMAN
|
||||||
| NC score | 0.019184 (rank : 90) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86UU1, O75133, Q4KMF8, Q8TEQ2 | Gene names | PHLDB1, DLNB07, KIAA0638, LL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
IRS2_MOUSE
|
||||||
| NC score | 0.018596 (rank : 91) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
FOXN1_HUMAN
|
||||||
| NC score | 0.018427 (rank : 92) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15353, O15352 | Gene names | FOXN1, WHN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude). | |||||
|
CXXC6_HUMAN
|
||||||
| NC score | 0.018131 (rank : 93) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
FBX46_MOUSE
|
||||||
| NC score | 0.017653 (rank : 94) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BG80 | Gene names | Fbxo46, Fbx46, Fbxo34l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
DAB2_HUMAN
|
||||||
| NC score | 0.017341 (rank : 95) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
XKR7_MOUSE
|
||||||
| NC score | 0.017309 (rank : 96) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5GH64 | Gene names | Xkr7, Xrg7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 7. | |||||
|
MTA2_HUMAN
|
||||||
| NC score | 0.017208 (rank : 97) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94776, Q9UQB5 | Gene names | MTA2, MTA1L1, PID | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1) (MTA1-L1 protein) (p53 target protein in deacetylase complex). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.017056 (rank : 98) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
BIN1_HUMAN
|
||||||
| NC score | 0.016711 (rank : 99) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
GRIN1_MOUSE
|
||||||
| NC score | 0.016352 (rank : 100) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
RFTN1_HUMAN
|
||||||
| NC score | 0.015830 (rank : 101) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14699, Q496Y2, Q4QQI7, Q5JB48, Q7Z7P2 | Gene names | RFTN1, KIAA0084 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Raftlin (Raft-linking protein) (Cell migration-inducing gene 2 protein). | |||||
|
BICD1_HUMAN
|
||||||
| NC score | 0.015786 (rank : 102) | θ value | 0.163984 (rank : 46) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
ADM2_MOUSE
|
||||||
| NC score | 0.015616 (rank : 103) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TNK8 | Gene names | Adm2, Am2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADM2 precursor (Intermedin) [Contains: Adrenomedullin-2 (Intermedin- long) (IMDL); Intermedin-short (IMDS)]. | |||||
|
BIN1_MOUSE
|
||||||
| NC score | 0.015052 (rank : 104) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08539, Q62434 | Gene names | Bin1, Amphl, Sh3p9 | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (SH3-domain-containing protein 9). | |||||
|
GA2L2_HUMAN
|
||||||
| NC score | 0.014554 (rank : 105) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
FOXN1_MOUSE
|
||||||
| NC score | 0.013901 (rank : 106) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
TENS4_HUMAN
|
||||||
| NC score | 0.013679 (rank : 107) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IZW8, Q71RB7, Q8WV64, Q96JV4 | Gene names | TNS4, CTEN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor (C-terminal tensin-like protein). | |||||
|
CASZ1_HUMAN
|
||||||
| NC score | 0.012811 (rank : 108) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 473 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86V15, Q5T9S1, Q8WX49, Q8WX50, Q9BT16, Q9NXC6 | Gene names | CASZ1, CST, ZNF693 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Castor homolog 1 zinc finger protein (Castor-related protein) (Zinc finger protein 693). | |||||
|
CMTA2_MOUSE
|
||||||
| NC score | 0.012360 (rank : 109) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80Y50, Q3TFK0, Q5SX68, Q80TP1, Q8R0D9, Q8R2N5 | Gene names | Camta2, Kiaa0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
BICD1_MOUSE
|
||||||
| NC score | 0.012342 (rank : 110) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
DCBD1_MOUSE
|
||||||
| NC score | 0.012200 (rank : 111) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D4J3, Q8R327, Q9D696 | Gene names | Dcbld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.012111 (rank : 112) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.011985 (rank : 113) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
SEM6C_MOUSE
|
||||||
| NC score | 0.011659 (rank : 114) | θ value | 0.62314 (rank : 54) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SI1L3_HUMAN
|
||||||
| NC score | 0.010683 (rank : 115) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
COIA1_HUMAN
|
||||||
| NC score | 0.010434 (rank : 116) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
PK2L1_HUMAN
|
||||||
| NC score | 0.010384 (rank : 117) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
HD_HUMAN
|
||||||
| NC score | 0.010248 (rank : 118) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.010043 (rank : 119) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.009776 (rank : 120) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.009683 (rank : 121) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
LIMD1_MOUSE
|
||||||
| NC score | 0.008561 (rank : 122) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
K0423_MOUSE
|
||||||
| NC score | 0.007873 (rank : 123) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6A070 | Gene names | Kiaa0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
RUNX1_MOUSE
|
||||||
| NC score | 0.007645 (rank : 124) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q03347, O08598, Q62049, Q9ESB9, Q9ET65 | Gene names | Runx1, Aml1, Cbfa2, Pebp2ab | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
WD21C_HUMAN
|
||||||
| NC score | 0.007372 (rank : 125) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NA75 | Gene names | WDR21C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 21C. | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.007320 (rank : 126) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.006896 (rank : 127) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
RBNS5_MOUSE
|
||||||
| NC score | 0.006702 (rank : 128) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80Y56, Q8K0L6, Q9CTW0 | Gene names | Zfyve20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20). | |||||
|
TNR4_HUMAN
|
||||||
| NC score | 0.006700 (rank : 129) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P43489, Q13663, Q5T7M0 | Gene names | TNFRSF4, TXGP1L | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 4 precursor (OX40L receptor) (ACT35 antigen) (TAX transcriptionally-activated glycoprotein 1 receptor) (CD134 antigen). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.006603 (rank : 130) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
TSC1_HUMAN
|
||||||
| NC score | 0.006245 (rank : 131) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92574 | Gene names | TSC1, KIAA0243, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Hamartin (Tuberous sclerosis 1 protein). | |||||
|
DUS2_HUMAN
|
||||||
| NC score | 0.005703 (rank : 132) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q05923 | Gene names | DUSP2, PAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.005123 (rank : 133) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
SLY_MOUSE
|
||||||
| NC score | 0.004987 (rank : 134) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K352, Q3TBX0, Q9DBV2 | Gene names | Sly | |||
|
Domain Architecture |

|
|||||
| Description | SH3 protein expressed in lymphocytes. | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.004752 (rank : 135) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
TBX2_HUMAN
|
||||||
| NC score | 0.004541 (rank : 136) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13207, Q16424, Q7Z647 | Gene names | TBX2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
ARHG3_HUMAN
|
||||||
| NC score | 0.002926 (rank : 137) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NR81, Q6NUN3, Q7Z4U2, Q7Z5T2, Q9H7T4 | Gene names | ARHGEF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 3 (Exchange factor found in platelets and leukemic and neuronal tissues) (XPLN). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.002808 (rank : 138) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
ATS13_HUMAN
|
||||||
| NC score | 0.002560 (rank : 139) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
HCLS1_MOUSE
|
||||||
| NC score | 0.001873 (rank : 140) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49710 | Gene names | Hcls1, Hs1 | |||
|
Domain Architecture |

|
|||||
| Description | Hematopoietic lineage cell-specific protein (Hematopoietic cell- specific LYN substrate 1) (LckBP1). | |||||
|
ZO2_MOUSE
|
||||||
| NC score | 0.001760 (rank : 141) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0U1, Q8K210 | Gene names | Tjp2, Zo2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
CAMKV_HUMAN
|
||||||
| NC score | 0.001534 (rank : 142) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
KIRR2_MOUSE
|
||||||
| NC score | 0.001501 (rank : 143) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TSU7, Q7TQ98 | Gene names | Kirrel2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kin of IRRE-like protein 2 precursor (Kin of irregular chiasm-like protein 2). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.000370 (rank : 144) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
NIN_MOUSE
|
||||||
| NC score | -0.000241 (rank : 145) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
ZN227_HUMAN
|
||||||
| NC score | -0.003956 (rank : 146) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 781 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86WZ6 | Gene names | ZNF227 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 227. | |||||
|
ZN169_HUMAN
|
||||||
| NC score | -0.004128 (rank : 147) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14929, Q6PI28 | Gene names | ZNF169 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 169. | |||||