Please be patient as the page loads
|
ADM2_MOUSE
|
||||||
| SwissProt Accessions | Q7TNK8 | Gene names | Adm2, Am2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADM2 precursor (Intermedin) [Contains: Adrenomedullin-2 (Intermedin- long) (IMDL); Intermedin-short (IMDS)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ADM2_MOUSE
|
||||||
| θ value | 8.20983e-86 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7TNK8 | Gene names | Adm2, Am2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADM2 precursor (Intermedin) [Contains: Adrenomedullin-2 (Intermedin- long) (IMDL); Intermedin-short (IMDS)]. | |||||
|
ADM2_HUMAN
|
||||||
| θ value | 1.19932e-44 (rank : 2) | NC score | 0.951761 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z4H4 | Gene names | ADM2, AM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADM2 precursor (Intermedin) [Contains: Adrenomedullin-2 (Intermedin- long) (IMDL); Intermedin-short (IMDS)]. | |||||
|
CA104_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 3) | NC score | 0.153496 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q66K80, Q8NA07 | Gene names | C1orf104 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf104. | |||||
|
ADML_HUMAN
|
||||||
| θ value | 0.21417 (rank : 4) | NC score | 0.182900 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35318, Q6FGW2 | Gene names | ADM, AM | |||
|
Domain Architecture |

|
|||||
| Description | ADM precursor [Contains: Adrenomedullin (AM); Proadrenomedullin N-20 terminal peptide (ProAM-N20) (ProAM N-terminal 20 peptide) (PAMP)]. | |||||
|
ASXL1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 5) | NC score | 0.072656 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IXJ9, Q5JWS9, Q8IYY7, Q9H466, Q9NQF8, Q9UFJ0, Q9UFP8, Q9Y2I4 | Gene names | ASXL1, KIAA0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
ARTN_HUMAN
|
||||||
| θ value | 0.47712 (rank : 6) | NC score | 0.069949 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T4W7, O95441, O96030, Q6P6A3 | Gene names | ARTN, EVN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Artemin precursor (Enovin) (Neublastin). | |||||
|
ADML_MOUSE
|
||||||
| θ value | 0.62314 (rank : 7) | NC score | 0.166600 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97297, P97453 | Gene names | Adm | |||
|
Domain Architecture |

|
|||||
| Description | ADM precursor [Contains: Adrenomedullin (AM); Proadrenomedullin N-20 terminal peptide (ProAM-N20) (ProAM N-terminal 20 peptide) (PAMP)]. | |||||
|
FBLI1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 8) | NC score | 0.026555 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WUP2, Q5VVE0, Q5VVE1, Q8IX23, Q96T00, Q9UFD6 | Gene names | FBLIM1, FBLP1 | |||
|
Domain Architecture |
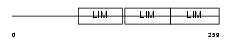
|
|||||
| Description | Filamin-binding LIM protein 1 (Filamin-binding LIM protein 1) (FBLP-1) (Mitogen-inducible 2-interacting protein) (MIG2-interacting protein) (Migfilin). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 1.38821 (rank : 9) | NC score | 0.033820 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 10) | NC score | 0.049191 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 1.81305 (rank : 11) | NC score | 0.043317 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
GLI3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 12) | NC score | 0.007913 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10071, O75219, Q9UDT5, Q9UJ39 | Gene names | GLI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | 2.36792 (rank : 13) | NC score | 0.032639 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.017742 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.014789 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
PRP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.026314 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
RERE_MOUSE
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.041617 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.027516 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.028787 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
KCNH3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.015616 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
5NT1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.024121 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96P26, Q8N9W3, Q8NA26, Q96DU5, Q96KE6, Q96M25, Q96SA3 | Gene names | NT5C1B, AIRP | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic 5'-nucleotidase 1B (EC 3.1.3.5) (Cytosolic 5'-nucleotidase IB) (cN1B) (cN-IB) (Autoimmune infertility-related protein). | |||||
|
CO3A1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.014879 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
HCN3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.013618 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
HIC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.002934 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14526 | Gene names | HIC1, ZBTB29 | |||
|
Domain Architecture |
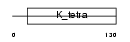
|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1) (Zinc finger and BTB domain-containing protein 29). | |||||
|
MICA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.013539 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94851, Q7Z3A8 | Gene names | MICAL2, KIAA0750 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
S39A5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.021425 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZMH5, Q8N6Y3 | Gene names | SLC39A5, ZIP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc transporter ZIP5 precursor (Solute carrier family 39 member 5). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.021012 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
BARH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.006814 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NY43, Q7Z4N7 | Gene names | BARHL2 | |||
|
Domain Architecture |

|
|||||
| Description | BarH-like 2 homeobox protein. | |||||
|
CENG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.008528 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99490, O00578, Q548E0, Q8IWU3 | Gene names | CENTG1, AGAP2, KIAA0167 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE) (GTP-binding and GTPase-activating protein 2) (GGAP2). | |||||
|
NID2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.006137 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88322 | Gene names | Nid2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Entactin-2). | |||||
|
ADM2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 8.20983e-86 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7TNK8 | Gene names | Adm2, Am2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADM2 precursor (Intermedin) [Contains: Adrenomedullin-2 (Intermedin- long) (IMDL); Intermedin-short (IMDS)]. | |||||
|
ADM2_HUMAN
|
||||||
| NC score | 0.951761 (rank : 2) | θ value | 1.19932e-44 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z4H4 | Gene names | ADM2, AM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADM2 precursor (Intermedin) [Contains: Adrenomedullin-2 (Intermedin- long) (IMDL); Intermedin-short (IMDS)]. | |||||
|
ADML_HUMAN
|
||||||
| NC score | 0.182900 (rank : 3) | θ value | 0.21417 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35318, Q6FGW2 | Gene names | ADM, AM | |||
|
Domain Architecture |

|
|||||
| Description | ADM precursor [Contains: Adrenomedullin (AM); Proadrenomedullin N-20 terminal peptide (ProAM-N20) (ProAM N-terminal 20 peptide) (PAMP)]. | |||||
|
ADML_MOUSE
|
||||||
| NC score | 0.166600 (rank : 4) | θ value | 0.62314 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97297, P97453 | Gene names | Adm | |||
|
Domain Architecture |

|
|||||
| Description | ADM precursor [Contains: Adrenomedullin (AM); Proadrenomedullin N-20 terminal peptide (ProAM-N20) (ProAM N-terminal 20 peptide) (PAMP)]. | |||||
|
CA104_HUMAN
|
||||||
| NC score | 0.153496 (rank : 5) | θ value | 0.0961366 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q66K80, Q8NA07 | Gene names | C1orf104 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf104. | |||||
|
ASXL1_HUMAN
|
||||||
| NC score | 0.072656 (rank : 6) | θ value | 0.365318 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IXJ9, Q5JWS9, Q8IYY7, Q9H466, Q9NQF8, Q9UFJ0, Q9UFP8, Q9Y2I4 | Gene names | ASXL1, KIAA0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
ARTN_HUMAN
|
||||||
| NC score | 0.069949 (rank : 7) | θ value | 0.47712 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T4W7, O95441, O96030, Q6P6A3 | Gene names | ARTN, EVN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Artemin precursor (Enovin) (Neublastin). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.049191 (rank : 8) | θ value | 1.38821 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.043317 (rank : 9) | θ value | 1.81305 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.041617 (rank : 10) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.033820 (rank : 11) | θ value | 1.38821 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.032639 (rank : 12) | θ value | 2.36792 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.028787 (rank : 13) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.027516 (rank : 14) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
FBLI1_HUMAN
|
||||||
| NC score | 0.026555 (rank : 15) | θ value | 1.06291 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WUP2, Q5VVE0, Q5VVE1, Q8IX23, Q96T00, Q9UFD6 | Gene names | FBLIM1, FBLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-binding LIM protein 1 (Filamin-binding LIM protein 1) (FBLP-1) (Mitogen-inducible 2-interacting protein) (MIG2-interacting protein) (Migfilin). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.026314 (rank : 16) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
5NT1B_HUMAN
|
||||||
| NC score | 0.024121 (rank : 17) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96P26, Q8N9W3, Q8NA26, Q96DU5, Q96KE6, Q96M25, Q96SA3 | Gene names | NT5C1B, AIRP | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic 5'-nucleotidase 1B (EC 3.1.3.5) (Cytosolic 5'-nucleotidase IB) (cN1B) (cN-IB) (Autoimmune infertility-related protein). | |||||
|
S39A5_HUMAN
|
||||||
| NC score | 0.021425 (rank : 18) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZMH5, Q8N6Y3 | Gene names | SLC39A5, ZIP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc transporter ZIP5 precursor (Solute carrier family 39 member 5). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.021012 (rank : 19) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.017742 (rank : 20) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
KCNH3_MOUSE
|
||||||
| NC score | 0.015616 (rank : 21) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
CO3A1_MOUSE
|
||||||
| NC score | 0.014879 (rank : 22) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.014789 (rank : 23) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
HCN3_MOUSE
|
||||||
| NC score | 0.013618 (rank : 24) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
MICA2_HUMAN
|
||||||
| NC score | 0.013539 (rank : 25) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94851, Q7Z3A8 | Gene names | MICAL2, KIAA0750 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
CENG1_HUMAN
|
||||||
| NC score | 0.008528 (rank : 26) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99490, O00578, Q548E0, Q8IWU3 | Gene names | CENTG1, AGAP2, KIAA0167 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE) (GTP-binding and GTPase-activating protein 2) (GGAP2). | |||||
|
GLI3_HUMAN
|
||||||
| NC score | 0.007913 (rank : 27) | θ value | 2.36792 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10071, O75219, Q9UDT5, Q9UJ39 | Gene names | GLI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
BARH2_HUMAN
|
||||||
| NC score | 0.006814 (rank : 28) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NY43, Q7Z4N7 | Gene names | BARHL2 | |||
|
Domain Architecture |

|
|||||
| Description | BarH-like 2 homeobox protein. | |||||
|
NID2_MOUSE
|
||||||
| NC score | 0.006137 (rank : 29) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88322 | Gene names | Nid2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Entactin-2). | |||||
|
HIC1_HUMAN
|
||||||
| NC score | 0.002934 (rank : 30) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14526 | Gene names | HIC1, ZBTB29 | |||
|
Domain Architecture |

|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1) (Zinc finger and BTB domain-containing protein 29). | |||||