Please be patient as the page loads
|
PK3CD_HUMAN
|
||||||
| SwissProt Accessions | O00329, O15445 | Gene names | PIK3CD | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PK3CA_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.981510 (rank : 6) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P42336, Q99762 | Gene names | PIK3CA | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
PK3CA_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.981681 (rank : 5) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P42337 | Gene names | Pik3ca | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
PK3CB_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.993666 (rank : 3) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42338 | Gene names | PIK3CB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CB_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.993613 (rank : 4) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BTI9 | Gene names | Pik3cb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CD_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00329, O15445 | Gene names | PIK3CD | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CD_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.999515 (rank : 2) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O35904 | Gene names | Pik3cd | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CG_MOUSE
|
||||||
| θ value | 1.52337e-140 (rank : 7) | NC score | 0.964122 (rank : 8) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JHG7 | Gene names | Pik3cg, Pi3kg1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
PK3CG_HUMAN
|
||||||
| θ value | 5.41814e-138 (rank : 8) | NC score | 0.964303 (rank : 7) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P48736, Q8IV23, Q9BZC8 | Gene names | PIK3CG | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
P3C2A_HUMAN
|
||||||
| θ value | 6.47494e-99 (rank : 9) | NC score | 0.824175 (rank : 14) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
P3C2B_HUMAN
|
||||||
| θ value | 7.91507e-97 (rank : 10) | NC score | 0.801978 (rank : 16) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00750, O95666, Q5SW99 | Gene names | PIK3C2B | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing beta polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-beta) (PtdIns-3-kinase C2 beta) (PI3K-C2beta) (C2-PI3K). | |||||
|
P3C2A_MOUSE
|
||||||
| θ value | 2.15548e-94 (rank : 11) | NC score | 0.825653 (rank : 13) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61194, Q61182 | Gene names | Pik3c2a, Cpk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha) (Cpk-m) (p170). | |||||
|
P3C2G_MOUSE
|
||||||
| θ value | 5.71191e-71 (rank : 12) | NC score | 0.893849 (rank : 10) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70167, O70168 | Gene names | Pik3c2g | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
P3C2G_HUMAN
|
||||||
| θ value | 2.3998e-69 (rank : 13) | NC score | 0.894318 (rank : 9) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75747 | Gene names | PIK3C2G | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
PK3C3_HUMAN
|
||||||
| θ value | 1.19932e-44 (rank : 14) | NC score | 0.866173 (rank : 11) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8NEB9, Q15134 | Gene names | PIK3C3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3) (Phosphatidylinositol 3-kinase p100 subunit). | |||||
|
PK3C3_MOUSE
|
||||||
| θ value | 1.56636e-44 (rank : 15) | NC score | 0.864925 (rank : 12) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6PF93, Q8R3S8 | Gene names | Pik3c3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3). | |||||
|
PI4KA_HUMAN
|
||||||
| θ value | 1.37858e-24 (rank : 16) | NC score | 0.804403 (rank : 15) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42356, Q7Z625, Q9UPG2 | Gene names | PIK4CA, PIK4 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase alpha (EC 2.7.1.67) (PI4-kinase alpha) (PtdIns-4-kinase alpha) (PI4K-alpha). | |||||
|
PI4KB_MOUSE
|
||||||
| θ value | 1.42661e-21 (rank : 17) | NC score | 0.767627 (rank : 17) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BKC8, Q8C146 | Gene names | Pik4cb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta). | |||||
|
PI4KB_HUMAN
|
||||||
| θ value | 5.42112e-21 (rank : 18) | NC score | 0.765207 (rank : 18) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UBF8, O15096, P78405, Q9BWR6 | Gene names | PIK4CB, PI4KB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta) (NPIK) (PI4K92). | |||||
|
PRKDC_HUMAN
|
||||||
| θ value | 9.26847e-13 (rank : 19) | NC score | 0.424664 (rank : 22) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
PRKDC_MOUSE
|
||||||
| θ value | 2.98157e-11 (rank : 20) | NC score | 0.428892 (rank : 21) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P97313, O88187, P97928, Q307W9, Q3V2W8, Q8C2A7, Q9Z341 | Gene names | Prkdc, Xrcc7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (P460). | |||||
|
FRAP_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 21) | NC score | 0.435269 (rank : 19) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JLN9, Q811J5, Q9CST1 | Gene names | Frap1, Frap | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
FRAP_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 22) | NC score | 0.433701 (rank : 20) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
ATM_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 23) | NC score | 0.365132 (rank : 23) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13315, O15429, Q12758, Q16551, Q93007, Q9NP02, Q9UCX7 | Gene names | ATM | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated) (A-T, mutated). | |||||
|
ATM_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 24) | NC score | 0.355535 (rank : 26) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62388 | Gene names | Atm | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated homolog) (A-T, mutated homolog). | |||||
|
ATR_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 25) | NC score | 0.358702 (rank : 24) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13535, Q7KYL3, Q93051, Q9BXK4 | Gene names | ATR, FRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein) (FRAP-related protein 1). | |||||
|
ATR_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 26) | NC score | 0.356409 (rank : 25) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JKK8 | Gene names | Atr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein). | |||||
|
SMG1_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 27) | NC score | 0.297705 (rank : 28) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
SMG1_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 28) | NC score | 0.300086 (rank : 27) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
TBLX_MOUSE
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.003568 (rank : 56) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QXE7, Q8BMM0, Q8BYQ4, Q8C0A1 | Gene names | Tbl1x, Tbl1 | |||
|
Domain Architecture |

|
|||||
| Description | F-box-like/WD repeat protein TBL1X (Transducin beta-like 1X protein). | |||||
|
DAB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.020336 (rank : 49) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
FA35A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.011802 (rank : 55) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3UEN2 | Gene names | Fam35a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM35A. | |||||
|
VPS18_MOUSE
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.016057 (rank : 51) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R307, Q8BGV6, Q8BZX6, Q8C126 | Gene names | Vps18 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 18. | |||||
|
MA2C1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.013841 (rank : 53) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NTJ4, Q13358, Q9UL64 | Gene names | MAN2C1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-mannosidase 2C1 (EC 3.2.1.24) (Alpha-D-mannoside mannohydrolase) (Mannosidase alpha class 2C member 1) (Alpha mannosidase 6A8B). | |||||
|
TBL1X_HUMAN
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.002889 (rank : 58) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60907, Q86UY2 | Gene names | TBL1X, TBL1 | |||
|
Domain Architecture |

|
|||||
| Description | F-box-like/WD repeat protein TBL1X (Transducin beta-like 1X protein) (Transducin-beta-like 1, X-linked) (SMAP55). | |||||
|
UDB11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.002791 (rank : 59) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75310, Q3KNV9 | Gene names | UGT2B11 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucuronosyltransferase 2B11 precursor (EC 2.4.1.17) (UDPGT). | |||||
|
ZRAB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.013182 (rank : 54) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95218, Q9UP63 | Gene names | ZRANB2, ZIS, ZNF265 | |||
|
Domain Architecture |
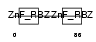
|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 2 Zinc finger protein 265 (Zinc finger, splicing). | |||||
|
BUP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.025434 (rank : 48) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UBR1, Q9UIR3 | Gene names | UPB1, BUP1 | |||
|
Domain Architecture |
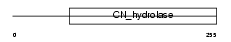
|
|||||
| Description | Beta-ureidopropionase (EC 3.5.1.6) (Beta-alanine synthase) (N- carbamoyl-beta-alanine amidohydrolase) (BUP-1). | |||||
|
DAB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.017320 (rank : 50) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97318, P97316, P97317 | Gene names | Dab1 | |||
|
Domain Architecture |
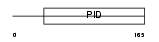
|
|||||
| Description | Disabled homolog 1. | |||||
|
SCN2A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.003119 (rank : 57) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
VPS18_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.014083 (rank : 52) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P253, Q8TCG0, Q96DI3, Q9H268 | Gene names | VPS18, KIAA1475 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 18 (hVPS18). | |||||
|
FA62A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.071794 (rank : 32) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BSJ8, O94848, Q6PJN4, Q9H6J1, Q9H6W2, Q9Y416 | Gene names | FAM62A, KIAA0747, MBC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
FA62A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.080498 (rank : 31) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3U7R1, Q8C8R1, Q91X62, Q9CVH0, Q9Z1X5, Q9Z1X6 | Gene names | Fam62a, Mbc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
RIMS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.066688 (rank : 34) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
RIMS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.056790 (rank : 42) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
RIMS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.056847 (rank : 41) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EQZ7, Q8C433, Q8CCK2 | Gene names | Rims2, Rab3ip2, Rim2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2) (Rab3-interacting protein 2). | |||||
|
RIMS3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.050287 (rank : 47) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UJD0, Q92511 | Gene names | RIMS3, KIAA0237 | |||
|
Domain Architecture |
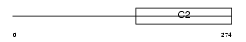
|
|||||
| Description | Regulating synaptic membrane exocytosis protein 3 (Nim3) (Rab-3- interacting molecule 3) (RIM 3) (RIM3 gamma). | |||||
|
RIMS4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.081095 (rank : 30) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H426, Q5JWT7 | Gene names | RIMS4 | |||
|
Domain Architecture |
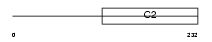
|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
RIMS4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.081098 (rank : 29) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P60191 | Gene names | Rims4 | |||
|
Domain Architecture |
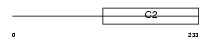
|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
SYTL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.060876 (rank : 37) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
SYTL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.057011 (rank : 40) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
SYTL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.054056 (rank : 45) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
SYTL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.052784 (rank : 46) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
SYTL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.071064 (rank : 33) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99N48, Q8C506, Q99N47, Q99N49, Q99N54, Q99N79 | Gene names | Sytl3, Slp3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 3 (Exophilin-6). | |||||
|
SYTL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.055142 (rank : 43) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96C24, Q5H9J3, Q5JPG8, Q8N9P4, Q9H4R0, Q9H4R1 | Gene names | SYTL4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
SYTL4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.054805 (rank : 44) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R0Q1, Q8R321, Q9R0Q0 | Gene names | Sytl4, Slp4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
SYTL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.059493 (rank : 38) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TDW5 | Gene names | SYTL5, SLP5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
SYTL5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.058943 (rank : 39) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80T23, Q812E5 | Gene names | Sytl5, Slp5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
TRRAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.063532 (rank : 35) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
TRRAP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.062723 (rank : 36) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80YV3, Q8C0Z5, Q8K104 | Gene names | Trrap | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (Tra1 homolog). | |||||
|
PK3CD_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00329, O15445 | Gene names | PIK3CD | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CD_MOUSE
|
||||||
| NC score | 0.999515 (rank : 2) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O35904 | Gene names | Pik3cd | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CB_HUMAN
|
||||||
| NC score | 0.993666 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42338 | Gene names | PIK3CB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CB_MOUSE
|
||||||
| NC score | 0.993613 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BTI9 | Gene names | Pik3cb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CA_MOUSE
|
||||||
| NC score | 0.981681 (rank : 5) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P42337 | Gene names | Pik3ca | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
PK3CA_HUMAN
|
||||||
| NC score | 0.981510 (rank : 6) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P42336, Q99762 | Gene names | PIK3CA | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
PK3CG_HUMAN
|
||||||
| NC score | 0.964303 (rank : 7) | θ value | 5.41814e-138 (rank : 8) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P48736, Q8IV23, Q9BZC8 | Gene names | PIK3CG | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
PK3CG_MOUSE
|
||||||
| NC score | 0.964122 (rank : 8) | θ value | 1.52337e-140 (rank : 7) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JHG7 | Gene names | Pik3cg, Pi3kg1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
P3C2G_HUMAN
|
||||||
| NC score | 0.894318 (rank : 9) | θ value | 2.3998e-69 (rank : 13) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75747 | Gene names | PIK3C2G | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
P3C2G_MOUSE
|
||||||
| NC score | 0.893849 (rank : 10) | θ value | 5.71191e-71 (rank : 12) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70167, O70168 | Gene names | Pik3c2g | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
PK3C3_HUMAN
|
||||||
| NC score | 0.866173 (rank : 11) | θ value | 1.19932e-44 (rank : 14) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8NEB9, Q15134 | Gene names | PIK3C3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3) (Phosphatidylinositol 3-kinase p100 subunit). | |||||
|
PK3C3_MOUSE
|
||||||
| NC score | 0.864925 (rank : 12) | θ value | 1.56636e-44 (rank : 15) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6PF93, Q8R3S8 | Gene names | Pik3c3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3). | |||||
|
P3C2A_MOUSE
|
||||||
| NC score | 0.825653 (rank : 13) | θ value | 2.15548e-94 (rank : 11) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61194, Q61182 | Gene names | Pik3c2a, Cpk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha) (Cpk-m) (p170). | |||||
|
P3C2A_HUMAN
|
||||||
| NC score | 0.824175 (rank : 14) | θ value | 6.47494e-99 (rank : 9) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
PI4KA_HUMAN
|
||||||
| NC score | 0.804403 (rank : 15) | θ value | 1.37858e-24 (rank : 16) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42356, Q7Z625, Q9UPG2 | Gene names | PIK4CA, PIK4 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase alpha (EC 2.7.1.67) (PI4-kinase alpha) (PtdIns-4-kinase alpha) (PI4K-alpha). | |||||
|
P3C2B_HUMAN
|
||||||
| NC score | 0.801978 (rank : 16) | θ value | 7.91507e-97 (rank : 10) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00750, O95666, Q5SW99 | Gene names | PIK3C2B | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing beta polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-beta) (PtdIns-3-kinase C2 beta) (PI3K-C2beta) (C2-PI3K). | |||||
|
PI4KB_MOUSE
|
||||||
| NC score | 0.767627 (rank : 17) | θ value | 1.42661e-21 (rank : 17) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BKC8, Q8C146 | Gene names | Pik4cb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta). | |||||
|
PI4KB_HUMAN
|
||||||
| NC score | 0.765207 (rank : 18) | θ value | 5.42112e-21 (rank : 18) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UBF8, O15096, P78405, Q9BWR6 | Gene names | PIK4CB, PI4KB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta) (NPIK) (PI4K92). | |||||
|
FRAP_MOUSE
|
||||||
| NC score | 0.435269 (rank : 19) | θ value | 3.29651e-10 (rank : 21) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JLN9, Q811J5, Q9CST1 | Gene names | Frap1, Frap | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
FRAP_HUMAN
|
||||||
| NC score | 0.433701 (rank : 20) | θ value | 5.62301e-10 (rank : 22) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
PRKDC_MOUSE
|
||||||
| NC score | 0.428892 (rank : 21) | θ value | 2.98157e-11 (rank : 20) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P97313, O88187, P97928, Q307W9, Q3V2W8, Q8C2A7, Q9Z341 | Gene names | Prkdc, Xrcc7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (P460). | |||||
|
PRKDC_HUMAN
|
||||||
| NC score | 0.424664 (rank : 22) | θ value | 9.26847e-13 (rank : 19) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
ATM_HUMAN
|
||||||
| NC score | 0.365132 (rank : 23) | θ value | 9.92553e-07 (rank : 23) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13315, O15429, Q12758, Q16551, Q93007, Q9NP02, Q9UCX7 | Gene names | ATM | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated) (A-T, mutated). | |||||
|
ATR_HUMAN
|
||||||
| NC score | 0.358702 (rank : 24) | θ value | 2.44474e-05 (rank : 25) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13535, Q7KYL3, Q93051, Q9BXK4 | Gene names | ATR, FRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein) (FRAP-related protein 1). | |||||
|
ATR_MOUSE
|
||||||
| NC score | 0.356409 (rank : 25) | θ value | 5.44631e-05 (rank : 26) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JKK8 | Gene names | Atr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein). | |||||
|
ATM_MOUSE
|
||||||
| NC score | 0.355535 (rank : 26) | θ value | 2.21117e-06 (rank : 24) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62388 | Gene names | Atm | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated homolog) (A-T, mutated homolog). | |||||
|
SMG1_MOUSE
|
||||||
| NC score | 0.300086 (rank : 27) | θ value | 0.00175202 (rank : 28) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
SMG1_HUMAN
|
||||||
| NC score | 0.297705 (rank : 28) | θ value | 0.00175202 (rank : 27) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
RIMS4_MOUSE
|
||||||
| NC score | 0.081098 (rank : 29) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P60191 | Gene names | Rims4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
RIMS4_HUMAN
|
||||||
| NC score | 0.081095 (rank : 30) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H426, Q5JWT7 | Gene names | RIMS4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
FA62A_MOUSE
|
||||||
| NC score | 0.080498 (rank : 31) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3U7R1, Q8C8R1, Q91X62, Q9CVH0, Q9Z1X5, Q9Z1X6 | Gene names | Fam62a, Mbc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
FA62A_HUMAN
|
||||||
| NC score | 0.071794 (rank : 32) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BSJ8, O94848, Q6PJN4, Q9H6J1, Q9H6W2, Q9Y416 | Gene names | FAM62A, KIAA0747, MBC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
SYTL3_MOUSE
|
||||||
| NC score | 0.071064 (rank : 33) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99N48, Q8C506, Q99N47, Q99N49, Q99N54, Q99N79 | Gene names | Sytl3, Slp3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 3 (Exophilin-6). | |||||
|
RIMS1_HUMAN
|
||||||
| NC score | 0.066688 (rank : 34) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
TRRAP_HUMAN
|
||||||
| NC score | 0.063532 (rank : 35) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
TRRAP_MOUSE
|
||||||
| NC score | 0.062723 (rank : 36) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80YV3, Q8C0Z5, Q8K104 | Gene names | Trrap | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (Tra1 homolog). | |||||
|
SYTL1_HUMAN
|
||||||
| NC score | 0.060876 (rank : 37) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
SYTL5_HUMAN
|
||||||
| NC score | 0.059493 (rank : 38) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TDW5 | Gene names | SYTL5, SLP5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
SYTL5_MOUSE
|
||||||
| NC score | 0.058943 (rank : 39) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80T23, Q812E5 | Gene names | Sytl5, Slp5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
SYTL1_MOUSE
|
||||||
| NC score | 0.057011 (rank : 40) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
RIMS2_MOUSE
|
||||||
| NC score | 0.056847 (rank : 41) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EQZ7, Q8C433, Q8CCK2 | Gene names | Rims2, Rab3ip2, Rim2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2) (Rab3-interacting protein 2). | |||||
|
RIMS2_HUMAN
|
||||||
| NC score | 0.056790 (rank : 42) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
SYTL4_HUMAN
|
||||||
| NC score | 0.055142 (rank : 43) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96C24, Q5H9J3, Q5JPG8, Q8N9P4, Q9H4R0, Q9H4R1 | Gene names | SYTL4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
SYTL4_MOUSE
|
||||||
| NC score | 0.054805 (rank : 44) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R0Q1, Q8R321, Q9R0Q0 | Gene names | Sytl4, Slp4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
SYTL2_HUMAN
|
||||||
| NC score | 0.054056 (rank : 45) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
SYTL2_MOUSE
|
||||||
| NC score | 0.052784 (rank : 46) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
RIMS3_HUMAN
|
||||||
| NC score | 0.050287 (rank : 47) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UJD0, Q92511 | Gene names | RIMS3, KIAA0237 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 3 (Nim3) (Rab-3- interacting molecule 3) (RIM 3) (RIM3 gamma). | |||||
|
BUP1_HUMAN
|
||||||
| NC score | 0.025434 (rank : 48) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UBR1, Q9UIR3 | Gene names | UPB1, BUP1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-ureidopropionase (EC 3.5.1.6) (Beta-alanine synthase) (N- carbamoyl-beta-alanine amidohydrolase) (BUP-1). | |||||
|
DAB1_HUMAN
|
||||||
| NC score | 0.020336 (rank : 49) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
DAB1_MOUSE
|
||||||
| NC score | 0.017320 (rank : 50) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97318, P97316, P97317 | Gene names | Dab1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
VPS18_MOUSE
|
||||||
| NC score | 0.016057 (rank : 51) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R307, Q8BGV6, Q8BZX6, Q8C126 | Gene names | Vps18 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 18. | |||||
|
VPS18_HUMAN
|
||||||
| NC score | 0.014083 (rank : 52) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P253, Q8TCG0, Q96DI3, Q9H268 | Gene names | VPS18, KIAA1475 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 18 (hVPS18). | |||||
|
MA2C1_HUMAN
|
||||||
| NC score | 0.013841 (rank : 53) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NTJ4, Q13358, Q9UL64 | Gene names | MAN2C1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-mannosidase 2C1 (EC 3.2.1.24) (Alpha-D-mannoside mannohydrolase) (Mannosidase alpha class 2C member 1) (Alpha mannosidase 6A8B). | |||||
|
ZRAB2_HUMAN
|
||||||
| NC score | 0.013182 (rank : 54) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95218, Q9UP63 | Gene names | ZRANB2, ZIS, ZNF265 | |||
|
Domain Architecture |
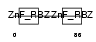
|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 2 Zinc finger protein 265 (Zinc finger, splicing). | |||||
|
FA35A_MOUSE
|
||||||
| NC score | 0.011802 (rank : 55) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3UEN2 | Gene names | Fam35a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM35A. | |||||
|
TBLX_MOUSE
|
||||||
| NC score | 0.003568 (rank : 56) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QXE7, Q8BMM0, Q8BYQ4, Q8C0A1 | Gene names | Tbl1x, Tbl1 | |||
|
Domain Architecture |

|
|||||
| Description | F-box-like/WD repeat protein TBL1X (Transducin beta-like 1X protein). | |||||
|
SCN2A_HUMAN
|
||||||
| NC score | 0.003119 (rank : 57) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
TBL1X_HUMAN
|
||||||
| NC score | 0.002889 (rank : 58) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60907, Q86UY2 | Gene names | TBL1X, TBL1 | |||
|
Domain Architecture |

|
|||||
| Description | F-box-like/WD repeat protein TBL1X (Transducin beta-like 1X protein) (Transducin-beta-like 1, X-linked) (SMAP55). | |||||
|
UDB11_HUMAN
|
||||||
| NC score | 0.002791 (rank : 59) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75310, Q3KNV9 | Gene names | UGT2B11 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucuronosyltransferase 2B11 precursor (EC 2.4.1.17) (UDPGT). | |||||