Please be patient as the page loads
|
RT15_MOUSE
|
||||||
| SwissProt Accessions | Q9DC71, Q7TMG9, Q8C7F1, Q9CVE0, Q9CWX1 | Gene names | Mrps15, Rpms15 | |||
|
Domain Architecture |
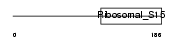
|
|||||
| Description | 28S ribosomal protein S15, mitochondrial precursor (S15mt) (MRP-S15). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RT15_MOUSE
|
||||||
| θ value | 7.35691e-119 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DC71, Q7TMG9, Q8C7F1, Q9CVE0, Q9CWX1 | Gene names | Mrps15, Rpms15 | |||
|
Domain Architecture |

|
|||||
| Description | 28S ribosomal protein S15, mitochondrial precursor (S15mt) (MRP-S15). | |||||
|
RT15_HUMAN
|
||||||
| θ value | 2.02684e-76 (rank : 2) | NC score | 0.930814 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P82914, Q9H2K1 | Gene names | MRPS15, RPMS15 | |||
|
Domain Architecture |

|
|||||
| Description | 28S ribosomal protein S15, mitochondrial precursor (S15mt) (MRP-S15). | |||||
|
TXLNG_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 3) | NC score | 0.103638 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NUQ3, Q9P0X1 | Gene names | TXLNG, CXorf15, LSR5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin (Lipopolysaccharide-specific response protein 5). | |||||
|
NEB2_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 4) | NC score | 0.113180 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
SDCG8_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 5) | NC score | 0.111348 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
NEB2_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 6) | NC score | 0.110491 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
SDCG8_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 7) | NC score | 0.099508 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
CI039_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 8) | NC score | 0.080277 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
TXLNG_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 9) | NC score | 0.104018 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BHN1, Q8BP11 | Gene names | Txlng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin. | |||||
|
UBCP1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 10) | NC score | 0.137963 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGR9, Q52L62, Q5XJH1, Q6PH96, Q99LT3 | Gene names | Ublcp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like domain-containing CTD phosphatase 1 (EC 3.1.3.16). | |||||
|
PLAP_HUMAN
|
||||||
| θ value | 0.125558 (rank : 11) | NC score | 0.052233 (rank : 61) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y263, Q53EU5, Q5VY33, Q9NUL8, Q9NVE9, Q9UF53, Q9Y5L1 | Gene names | PLAA, PLAP | |||
|
Domain Architecture |
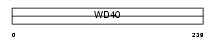
|
|||||
| Description | Phospholipase A-2-activating protein (PLAP) (PLA2P). | |||||
|
UBCP1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.134436 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WVY7, Q96DK5 | Gene names | UBLCP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like domain-containing CTD phosphatase 1 (EC 3.1.3.16). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.076745 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
SMC4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.084042 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.041379 (rank : 92) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
STMN1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.082529 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
STMN1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.081247 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.066873 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
LUZP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.079894 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.060750 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
K1967_HUMAN
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.048321 (rank : 90) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N163, Q6P0Q9, Q8N3G7, Q8N8M1, Q8TF34, Q9H9Q9, Q9HD12, Q9NT55 | Gene names | KIAA1967, DBC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1967 (Deleted in breast cancer gene 1 protein) (DBC.1) (DBC-1) (p30 DBC). | |||||
|
LUZP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.081386 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
PLAP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.045132 (rank : 91) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27612, Q8C6C4 | Gene names | Plaa, Plap | |||
|
Domain Architecture |
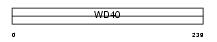
|
|||||
| Description | Phospholipase A-2-activating protein (PLAP). | |||||
|
RABE1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.060494 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.065009 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
SMC4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.079652 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CG47, Q8BTS7, Q8BTY9, Q99K21 | Gene names | Smc4l1, Capc, Smc4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (XCAP-C homolog). | |||||
|
CEP57_MOUSE
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.058842 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 580 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8CEE0, Q6ZQJ3, Q7TN18, Q80X65, Q810F2, Q9D4J4, Q9D5S4, Q9D5W5 | Gene names | Cep57, Kiaa0092, Tsp57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin). | |||||
|
CR034_HUMAN
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.058466 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5BJE1, Q6ZP67, Q6ZU20 | Gene names | C18orf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34. | |||||
|
IFT74_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.071819 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.069831 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.039876 (rank : 93) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.069354 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
TRI17_MOUSE
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.015470 (rank : 98) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 620 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TPM3, Q99PP8 | Gene names | Trim17 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 17. | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.059035 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CTCF_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.004274 (rank : 101) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49711 | Gene names | CTCF | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor CTCF (CCCTC-binding factor) (CTCFL paralog) (11-zinc finger protein). | |||||
|
CTCF_MOUSE
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.004204 (rank : 102) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61164 | Gene names | Ctcf | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor CTCF (CCCTC-binding factor) (CTCFL paralog) (11-zinc finger protein). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.053000 (rank : 57) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
SPAG5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.054835 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96R06, O95213, Q9BWE8, Q9NT17, Q9UFE6 | Gene names | SPAG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Astrin) (Mitotic spindle-associated protein p126) (MAP126) (Deepest). | |||||
|
STMN4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.056372 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H169 | Gene names | STMN4 | |||
|
Domain Architecture |
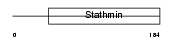
|
|||||
| Description | Stathmin-4 (Stathmin-like protein B3) (RB3). | |||||
|
STMN4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.056359 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P63042, O35414, O35415, O35416 | Gene names | Stmn4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stathmin-4 (Stathmin-like protein B3) (RB3). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.026131 (rank : 96) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
CCD41_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.056017 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
DMD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.038839 (rank : 94) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
PEPL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.051359 (rank : 68) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
PRS8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.013125 (rank : 99) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P62195, O35051, O43208, P47210, P52915, P52916 | Gene names | PSMC5, SUG1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (Thyroid hormone receptor- interacting protein 1) (TRIP1). | |||||
|
PRS8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.013125 (rank : 100) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P62196, O35051, P47210, P52915, P52916, Q3UL51, Q9CWN5 | Gene names | Psmc5, Sug1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (mSUG1). | |||||
|
ZSWM3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.019970 (rank : 97) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96MP5, Q9BR13 | Gene names | ZSWIM3, C20orf164 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 3. | |||||
|
BORIS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.003956 (rank : 103) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NI51, Q5JUG4, Q9BZ30, Q9NQJ3 | Gene names | CTCFL, BORIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor CTCFL (CCCTC-binding factor) (Brother of the regulator of imprinted sites) (Zinc finger protein CTCF-T) (CTCF paralog). | |||||
|
CTTB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.034515 (rank : 95) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
FYCO1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.049742 (rank : 89) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
CCD21_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.050068 (rank : 85) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CCD39_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.050868 (rank : 76) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CCD46_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.051428 (rank : 66) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CCD46_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.050301 (rank : 83) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CCD62_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.050864 (rank : 77) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6P9F0, Q6ZVF2, Q86VJ0, Q9BYZ5 | Gene names | CCDC62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 60 (Aaa-protein) (Protein TSP- NY). | |||||
|
CE290_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.054219 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CEP55_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.051423 (rank : 67) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
CF152_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.051759 (rank : 64) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
CI093_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.050717 (rank : 78) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
CN145_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.052829 (rank : 58) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
CP135_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.050906 (rank : 75) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CR034_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.059405 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CDV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34 homolog. | |||||
|
CY15A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.075338 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BZA5 | Gene names | CYorf15A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative testis protein CYorf15A. | |||||
|
EEA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.052326 (rank : 60) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.050384 (rank : 82) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.070644 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.050045 (rank : 88) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.050136 (rank : 84) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.050049 (rank : 87) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.052619 (rank : 59) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
IFT74_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.056823 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
IFT81_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.051693 (rank : 65) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
INCE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.050666 (rank : 79) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
INCE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.051050 (rank : 72) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
KMHN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.054183 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
KMHN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.053559 (rank : 52) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
MD1L1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.050062 (rank : 86) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
NEB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.079175 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
PEPL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.051144 (rank : 71) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
RABE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.054742 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.057648 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.051927 (rank : 63) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.050533 (rank : 81) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
SAS6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.053063 (rank : 56) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6UVJ0, Q8N3K0 | Gene names | SASS6, SAS6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog (HsSAS-6). | |||||
|
SAS6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.053383 (rank : 54) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
SM1L2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.068808 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
SM1L2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.067078 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.060719 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
SMC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.053796 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
SMC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.060558 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
SMC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.060571 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
STMN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.051249 (rank : 69) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q93045, O14952, Q6PK68 | Gene names | STMN2, SCG10, SCGN10 | |||
|
Domain Architecture |
|
|||||
| Description | Stathmin-2 (SCG10 protein) (Superior cervical ganglion-10 protein). | |||||
|
STMN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.051249 (rank : 70) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P55821 | Gene names | Stmn2, Scg10, Scgn10, Stmb2 | |||
|
Domain Architecture |
|
|||||
| Description | Stathmin-2 (SCG10 protein) (Superior cervical ganglion-10 protein). | |||||
|
TAXB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.052057 (rank : 62) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
TAXB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.053251 (rank : 55) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q3UKC1, Q91YT6, Q9CVF0, Q9DC45 | Gene names | Tax1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 homolog. | |||||
|
TPM3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.050999 (rank : 73) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P06753, P12324, Q969Q2, Q9NQH8 | Gene names | TPM3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma) (hTM5). | |||||
|
TPM3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.050951 (rank : 74) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P21107, Q09021, Q60606, Q80SW6, Q9EPW3 | Gene names | Tpm3, Tpm-5, Tpm5 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.050538 (rank : 80) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
TXLNA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.073047 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
TXLNA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.074844 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.072466 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
TXLNB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.079338 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8VBT1, Q3UVB8, Q8BUK2 | Gene names | Txlnb, Mdp77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77). | |||||
|
XE7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.053394 (rank : 53) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
RT15_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 7.35691e-119 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DC71, Q7TMG9, Q8C7F1, Q9CVE0, Q9CWX1 | Gene names | Mrps15, Rpms15 | |||
|
Domain Architecture |

|
|||||
| Description | 28S ribosomal protein S15, mitochondrial precursor (S15mt) (MRP-S15). | |||||
|
RT15_HUMAN
|
||||||
| NC score | 0.930814 (rank : 2) | θ value | 2.02684e-76 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P82914, Q9H2K1 | Gene names | MRPS15, RPMS15 | |||
|
Domain Architecture |

|
|||||
| Description | 28S ribosomal protein S15, mitochondrial precursor (S15mt) (MRP-S15). | |||||
|
UBCP1_MOUSE
|
||||||
| NC score | 0.137963 (rank : 3) | θ value | 0.0736092 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGR9, Q52L62, Q5XJH1, Q6PH96, Q99LT3 | Gene names | Ublcp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like domain-containing CTD phosphatase 1 (EC 3.1.3.16). | |||||
|
UBCP1_HUMAN
|
||||||
| NC score | 0.134436 (rank : 4) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WVY7, Q96DK5 | Gene names | UBLCP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like domain-containing CTD phosphatase 1 (EC 3.1.3.16). | |||||
|
NEB2_MOUSE
|
||||||
| NC score | 0.113180 (rank : 5) | θ value | 0.00869519 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
SDCG8_MOUSE
|
||||||
| NC score | 0.111348 (rank : 6) | θ value | 0.00869519 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
NEB2_HUMAN
|
||||||
| NC score | 0.110491 (rank : 7) | θ value | 0.0148317 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
TXLNG_MOUSE
|
||||||
| NC score | 0.104018 (rank : 8) | θ value | 0.0736092 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BHN1, Q8BP11 | Gene names | Txlng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin. | |||||
|
TXLNG_HUMAN
|
||||||
| NC score | 0.103638 (rank : 9) | θ value | 0.00509761 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NUQ3, Q9P0X1 | Gene names | TXLNG, CXorf15, LSR5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin (Lipopolysaccharide-specific response protein 5). | |||||
|
SDCG8_HUMAN
|
||||||
| NC score | 0.099508 (rank : 10) | θ value | 0.0193708 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
SMC4_HUMAN
|
||||||
| NC score | 0.084042 (rank : 11) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
STMN1_HUMAN
|
||||||
| NC score | 0.082529 (rank : 12) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
LUZP1_HUMAN
|
||||||
| NC score | 0.081386 (rank : 13) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
STMN1_MOUSE
|
||||||
| NC score | 0.081247 (rank : 14) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.080277 (rank : 15) | θ value | 0.0736092 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
LUZP1_MOUSE
|
||||||
| NC score | 0.079894 (rank : 16) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
SMC4_MOUSE
|
||||||
| NC score | 0.079652 (rank : 17) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CG47, Q8BTS7, Q8BTY9, Q99K21 | Gene names | Smc4l1, Capc, Smc4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (XCAP-C homolog). | |||||
|
TXLNB_MOUSE
|
||||||
| NC score | 0.079338 (rank : 18) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8VBT1, Q3UVB8, Q8BUK2 | Gene names | Txlnb, Mdp77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77). | |||||
|
NEB1_HUMAN
|
||||||
| NC score | 0.079175 (rank : 19) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.076745 (rank : 20) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
CY15A_HUMAN
|
||||||
| NC score | 0.075338 (rank : 21) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BZA5 | Gene names | CYorf15A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative testis protein CYorf15A. | |||||
|
TXLNA_MOUSE
|
||||||
| NC score | 0.074844 (rank : 22) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
TXLNA_HUMAN
|
||||||
| NC score | 0.073047 (rank : 23) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.072466 (rank : 24) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
IFT74_HUMAN
|
||||||
| NC score | 0.071819 (rank : 25) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.070644 (rank : 26) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.069831 (rank : 27) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.069354 (rank : 28) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SM1L2_HUMAN
|
||||||
| NC score | 0.068808 (rank : 29) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
SM1L2_MOUSE
|
||||||
| NC score | 0.067078 (rank : 30) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.066873 (rank : 31) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.065009 (rank : 32) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.060750 (rank : 33) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.060719 (rank : 34) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
SMC3_MOUSE
|
||||||
| NC score | 0.060571 (rank : 35) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
SMC3_HUMAN
|
||||||
| NC score | 0.060558 (rank : 36) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
RABE1_HUMAN
|
||||||
| NC score | 0.060494 (rank : 37) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
CR034_MOUSE
|
||||||
| NC score | 0.059405 (rank : 38) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CDV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34 homolog. | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.059035 (rank : 39) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CEP57_MOUSE
|
||||||
| NC score | 0.058842 (rank : 40) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 580 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8CEE0, Q6ZQJ3, Q7TN18, Q80X65, Q810F2, Q9D4J4, Q9D5S4, Q9D5W5 | Gene names | Cep57, Kiaa0092, Tsp57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin). | |||||
|
CR034_HUMAN
|
||||||
| NC score | 0.058466 (rank : 41) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5BJE1, Q6ZP67, Q6ZU20 | Gene names | C18orf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34. | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.057648 (rank : 42) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
IFT74_MOUSE
|
||||||
| NC score | 0.056823 (rank : 43) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
STMN4_HUMAN
|
||||||
| NC score | 0.056372 (rank : 44) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H169 | Gene names | STMN4 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin-4 (Stathmin-like protein B3) (RB3). | |||||
|
STMN4_MOUSE
|
||||||
| NC score | 0.056359 (rank : 45) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P63042, O35414, O35415, O35416 | Gene names | Stmn4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stathmin-4 (Stathmin-like protein B3) (RB3). | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.056017 (rank : 46) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
SPAG5_HUMAN
|
||||||
| NC score | 0.054835 (rank : 47) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96R06, O95213, Q9BWE8, Q9NT17, Q9UFE6 | Gene names | SPAG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Astrin) (Mitotic spindle-associated protein p126) (MAP126) (Deepest). | |||||
|
RABE1_MOUSE
|
||||||
| NC score | 0.054742 (rank : 48) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.054219 (rank : 49) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
KMHN1_HUMAN
|
||||||
| NC score | 0.054183 (rank : 50) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
SMC2_MOUSE
|
||||||
| NC score | 0.053796 (rank : 51) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
KMHN1_MOUSE
|
||||||
| NC score | 0.053559 (rank : 52) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
XE7_HUMAN
|
||||||
| NC score | 0.053394 (rank : 53) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
SAS6_MOUSE
|
||||||
| NC score | 0.053383 (rank : 54) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
TAXB1_MOUSE
|
||||||
| NC score | 0.053251 (rank : 55) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q3UKC1, Q91YT6, Q9CVF0, Q9DC45 | Gene names | Tax1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 homolog. | |||||
|
SAS6_HUMAN
|
||||||
| NC score | 0.053063 (rank : 56) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6UVJ0, Q8N3K0 | Gene names | SASS6, SAS6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog (HsSAS-6). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.053000 (rank : 57) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.052829 (rank : 58) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.052619 (rank : 59) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.052326 (rank : 60) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
PLAP_HUMAN
|
||||||
| NC score | 0.052233 (rank : 61) | θ value | 0.125558 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y263, Q53EU5, Q5VY33, Q9NUL8, Q9NVE9, Q9UF53, Q9Y5L1 | Gene names | PLAA, PLAP | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A-2-activating protein (PLAP) (PLA2P). | |||||
|
TAXB1_HUMAN
|
||||||
| NC score | 0.052057 (rank : 62) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.051927 (rank : 63) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
CF152_HUMAN
|
||||||
| NC score | 0.051759 (rank : 64) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
IFT81_MOUSE
|
||||||
| NC score | 0.051693 (rank : 65) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.051428 (rank : 66) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CEP55_MOUSE
|
||||||
| NC score | 0.051423 (rank : 67) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.051359 (rank : 68) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
STMN2_HUMAN
|
||||||
| NC score | 0.051249 (rank : 69) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q93045, O14952, Q6PK68 | Gene names | STMN2, SCG10, SCGN10 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin-2 (SCG10 protein) (Superior cervical ganglion-10 protein). | |||||
|
STMN2_MOUSE
|
||||||
| NC score | 0.051249 (rank : 70) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P55821 | Gene names | Stmn2, Scg10, Scgn10, Stmb2 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin-2 (SCG10 protein) (Superior cervical ganglion-10 protein). | |||||
|
PEPL_MOUSE
|
||||||
| NC score | 0.051144 (rank : 71) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.051050 (rank : 72) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
TPM3_HUMAN
|
||||||
| NC score | 0.050999 (rank : 73) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P06753, P12324, Q969Q2, Q9NQH8 | Gene names | TPM3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma) (hTM5). | |||||
|
TPM3_MOUSE
|
||||||
| NC score | 0.050951 (rank : 74) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P21107, Q09021, Q60606, Q80SW6, Q9EPW3 | Gene names | Tpm3, Tpm-5, Tpm5 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.050906 (rank : 75) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CCD39_MOUSE
|
||||||
| NC score | 0.050868 (rank : 76) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CCD62_HUMAN
|
||||||
| NC score | 0.050864 (rank : 77) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6P9F0, Q6ZVF2, Q86VJ0, Q9BYZ5 | Gene names | CCDC62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 60 (Aaa-protein) (Protein TSP- NY). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.050717 (rank : 78) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.050666 (rank : 79) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.050538 (rank : 80) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.050533 (rank : 81) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.050384 (rank : 82) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.050301 (rank : 83) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | 0.050136 (rank : 84) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.050068 (rank : 85) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
MD1L1_MOUSE
|
||||||
| NC score | 0.050062 (rank : 86) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.050049 (rank : 87) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.050045 (rank : 88) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
FYCO1_MOUSE
|
||||||
| NC score | 0.049742 (rank : 89) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
K1967_HUMAN
|
||||||
| NC score | 0.048321 (rank : 90) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N163, Q6P0Q9, Q8N3G7, Q8N8M1, Q8TF34, Q9H9Q9, Q9HD12, Q9NT55 | Gene names | KIAA1967, DBC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1967 (Deleted in breast cancer gene 1 protein) (DBC.1) (DBC-1) (p30 DBC). | |||||
|
PLAP_MOUSE
|
||||||
| NC score | 0.045132 (rank : 91) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27612, Q8C6C4 | Gene names | Plaa, Plap | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A-2-activating protein (PLAP). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.041379 (rank : 92) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.039876 (rank : 93) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.038839 (rank : 94) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
CTTB2_HUMAN
|
||||||
| NC score | 0.034515 (rank : 95) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.026131 (rank : 96) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
ZSWM3_HUMAN
|
||||||
| NC score | 0.019970 (rank : 97) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96MP5, Q9BR13 | Gene names | ZSWIM3, C20orf164 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 3. | |||||
|
TRI17_MOUSE
|
||||||
| NC score | 0.015470 (rank : 98) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 620 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TPM3, Q99PP8 | Gene names | Trim17 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 17. | |||||
|
PRS8_HUMAN
|
||||||
| NC score | 0.013125 (rank : 99) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P62195, O35051, O43208, P47210, P52915, P52916 | Gene names | PSMC5, SUG1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (Thyroid hormone receptor- interacting protein 1) (TRIP1). | |||||
|
PRS8_MOUSE
|
||||||
| NC score | 0.013125 (rank : 100) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P62196, O35051, P47210, P52915, P52916, Q3UL51, Q9CWN5 | Gene names | Psmc5, Sug1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 8 (Proteasome 26S subunit ATPase 5) (Proteasome subunit p45) (p45/SUG) (mSUG1). | |||||
|
CTCF_HUMAN
|
||||||
| NC score | 0.004274 (rank : 101) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49711 | Gene names | CTCF | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor CTCF (CCCTC-binding factor) (CTCFL paralog) (11-zinc finger protein). | |||||
|
CTCF_MOUSE
|
||||||
| NC score | 0.004204 (rank : 102) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61164 | Gene names | Ctcf | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor CTCF (CCCTC-binding factor) (CTCFL paralog) (11-zinc finger protein). | |||||
|
BORIS_HUMAN
|
||||||
| NC score | 0.003956 (rank : 103) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NI51, Q5JUG4, Q9BZ30, Q9NQJ3 | Gene names | CTCFL, BORIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor CTCFL (CCCTC-binding factor) (Brother of the regulator of imprinted sites) (Zinc finger protein CTCF-T) (CTCF paralog). | |||||