Please be patient as the page loads
|
NMDE1_MOUSE
|
||||||
| SwissProt Accessions | P35436 | Gene names | Grin2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NMDE1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998225 (rank : 2) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q12879, O00669 | Gene names | GRIN2A | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A) (hNR2A). | |||||
|
NMDE1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P35436 | Gene names | Grin2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A). | |||||
|
NMDE2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.992656 (rank : 3) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13224, Q12919, Q13220, Q13225, Q9UM56 | Gene names | GRIN2B | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B) (N-methyl-D-aspartate receptor subunit 3) (NR3) (hNR3). | |||||
|
NMDE2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.992493 (rank : 4) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q01097, Q9DCB2 | Gene names | Grin2b | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B). | |||||
|
NMDE3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.987121 (rank : 6) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.987585 (rank : 5) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q01098 | Gene names | Grin2c | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE4_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.984920 (rank : 7) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
|
NMDE4_MOUSE
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.982499 (rank : 8) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q03391 | Gene names | Grin2d | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D). | |||||
|
NMD3A_HUMAN
|
||||||
| θ value | 2.47191e-82 (rank : 9) | NC score | 0.933847 (rank : 9) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TCU5, Q8TF29, Q8WXI6 | Gene names | GRIN3A, KIAA1973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3A precursor (N-methyl-D-aspartate receptor subtype NR3A) (NMDAR-L). | |||||
|
NMDZ1_HUMAN
|
||||||
| θ value | 2.0926e-81 (rank : 10) | NC score | 0.908337 (rank : 12) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q05586, P35437, Q12867, Q12868 | Gene names | GRIN1, NMDAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMDZ1_MOUSE
|
||||||
| θ value | 4.66183e-81 (rank : 11) | NC score | 0.907859 (rank : 13) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35438, Q8CFS4 | Gene names | Grin1, Glurz1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMD3B_MOUSE
|
||||||
| θ value | 7.45998e-71 (rank : 12) | NC score | 0.925136 (rank : 10) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91ZU9, Q8VHV0 | Gene names | Grin3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subunit NR3B) (NR3B) (NMDAR3B) (NMDA receptor NR4) (Nr4). | |||||
|
NMD3B_HUMAN
|
||||||
| θ value | 5.00389e-67 (rank : 13) | NC score | 0.923653 (rank : 11) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60391, Q5EAK7, Q7RTW9 | Gene names | GRIN3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subtype NR3B) (NR3B) (NMDAR3B). | |||||
|
GRIK2_HUMAN
|
||||||
| θ value | 1.51363e-47 (rank : 14) | NC score | 0.836174 (rank : 22) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13002 | Gene names | GRIK2, GLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 2 precursor (Glutamate receptor 6) (GluR-6) (GluR6) (Excitatory amino acid receptor 4) (EAA4). | |||||
|
GRIK3_HUMAN
|
||||||
| θ value | 7.51208e-47 (rank : 15) | NC score | 0.838284 (rank : 17) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13003, Q13004, Q16136 | Gene names | GRIK3, GLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 3 precursor (Glutamate receptor 7) (GluR-7) (GluR7) (Excitatory amino acid receptor 5) (EAA5). | |||||
|
GRIK1_HUMAN
|
||||||
| θ value | 3.72821e-46 (rank : 16) | NC score | 0.837510 (rank : 19) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P39086, Q13001, Q86SU9 | Gene names | GRIK1, GLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5) (Excitatory amino acid receptor 3) (EAA3). | |||||
|
GRID2_MOUSE
|
||||||
| θ value | 2.41655e-45 (rank : 17) | NC score | 0.843840 (rank : 14) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61625 | Gene names | Grid2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRIA1_MOUSE
|
||||||
| θ value | 7.03111e-45 (rank : 18) | NC score | 0.830701 (rank : 25) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23818 | Gene names | Gria1, Glur1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIK5_HUMAN
|
||||||
| θ value | 7.03111e-45 (rank : 19) | NC score | 0.836080 (rank : 23) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16478 | Gene names | GRIK5, GRIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Excitatory amino acid receptor 2) (EAA2). | |||||
|
GRIK5_MOUSE
|
||||||
| θ value | 1.19932e-44 (rank : 20) | NC score | 0.835594 (rank : 24) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61626 | Gene names | Grik5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Glutamate receptor gamma-2) (GluR gamma-2). | |||||
|
GRIA1_HUMAN
|
||||||
| θ value | 2.04573e-44 (rank : 21) | NC score | 0.829335 (rank : 26) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42261 | Gene names | GRIA1, GLUH1, GLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIA3_HUMAN
|
||||||
| θ value | 2.67181e-44 (rank : 22) | NC score | 0.825308 (rank : 28) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42263, Q4VXD5, Q4VXD6, Q9HDA0, Q9HDA1, Q9HDA2 | Gene names | GRIA3, GLUR3, GLURC | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRIA3_MOUSE
|
||||||
| θ value | 4.55743e-44 (rank : 23) | NC score | 0.825288 (rank : 30) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2W9, Q5DTJ0 | Gene names | Gria3, Glur3, Kiaa4184 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRID2_HUMAN
|
||||||
| θ value | 5.95217e-44 (rank : 24) | NC score | 0.842495 (rank : 15) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43424 | Gene names | GRID2, GLURD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRIK4_HUMAN
|
||||||
| θ value | 1.32601e-43 (rank : 25) | NC score | 0.836907 (rank : 20) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16099 | Gene names | GRIK4, GRIK | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor (Glutamate receptor KA-1) (KA1) (Excitatory amino acid receptor 1) (EAA1). | |||||
|
GRIK4_MOUSE
|
||||||
| θ value | 1.32601e-43 (rank : 26) | NC score | 0.836663 (rank : 21) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BMF5 | Gene names | Grik4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor. | |||||
|
GRIA4_HUMAN
|
||||||
| θ value | 8.59492e-43 (rank : 27) | NC score | 0.827156 (rank : 27) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48058 | Gene names | GRIA4, GLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIA2_HUMAN
|
||||||
| θ value | 2.50074e-42 (rank : 28) | NC score | 0.825307 (rank : 29) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42262, Q96FP6 | Gene names | GRIA2, GLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIA2_MOUSE
|
||||||
| θ value | 4.2656e-42 (rank : 29) | NC score | 0.824644 (rank : 32) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23819, Q61604, Q61605, Q8BG69, Q8BXU3, Q9D6D3 | Gene names | Gria2, Glur2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIA4_MOUSE
|
||||||
| θ value | 9.50281e-42 (rank : 30) | NC score | 0.825008 (rank : 31) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2W8 | Gene names | Gria4, Glur4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRID1_MOUSE
|
||||||
| θ value | 1.79215e-40 (rank : 31) | NC score | 0.839376 (rank : 16) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61627 | Gene names | Grid1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRID1_HUMAN
|
||||||
| θ value | 8.89434e-40 (rank : 32) | NC score | 0.837835 (rank : 18) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULK0, Q8IXT3 | Gene names | GRID1, KIAA1220 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRIK1_MOUSE
|
||||||
| θ value | 1.37539e-32 (rank : 33) | NC score | 0.818341 (rank : 33) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q60934 | Gene names | Grik1, Glur5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5). | |||||
|
DLEC1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 34) | NC score | 0.054166 (rank : 35) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BLA1 | Gene names | Dlec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in lung and esophageal cancer protein 1 homolog. | |||||
|
ERB1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 35) | NC score | 0.139982 (rank : 34) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60509 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-R(b)_3p24.3 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-R(b) Env protein) [Includes: Surface protein domain (SU); Transmembrane protein domain (TM)]. | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 36) | NC score | 0.015338 (rank : 45) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 0.125558 (rank : 37) | NC score | 0.025394 (rank : 39) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
ERCC6_HUMAN
|
||||||
| θ value | 0.279714 (rank : 38) | NC score | 0.016183 (rank : 44) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03468 | Gene names | ERCC6, CSB | |||
|
Domain Architecture |

|
|||||
| Description | DNA excision repair protein ERCC-6 (EC 3.6.1.-) (ATP-dependent helicase ERCC6) (Cockayne syndrome protein CSB). | |||||
|
SFR12_MOUSE
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.021089 (rank : 40) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.011934 (rank : 49) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
CKAP2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.017354 (rank : 43) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WWK9, Q3KRA5, Q5VXB4, Q8IWV5, Q8IWV6, Q96FH9, Q9H012, Q9H0D0, Q9H988, Q9HC49, Q9NVG4 | Gene names | CKAP2, LB1, TMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 2 (Tumor-associated microtubule- associated protein) (CTCL tumor antigen se20-10). | |||||
|
GALT5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.006660 (rank : 64) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z7M9, Q9UGK7, Q9UHL6 | Gene names | GALNT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 5) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 5) (Polypeptide GalNAc transferase 5) (GalNAc-T5) (pp-GaNTase 5). | |||||
|
S19A3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.020137 (rank : 41) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99PL8, Q32MF5 | Gene names | Slc19a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thiamine transporter 2 (ThTr-2) (ThTr2) (Solute carrier family 19 member 3). | |||||
|
WNK3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.001615 (rank : 76) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYP7, Q9HCK6 | Gene names | WNK3, KIAA1566, PRKWNK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK3 (EC 2.7.11.1) (Protein kinase with no lysine 3) (Protein kinase, lysine-deficient 3). | |||||
|
CIR_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.029361 (rank : 37) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
PC11X_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.001644 (rank : 75) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZA7, Q2TJH0, Q2TJH1, Q2TJH3, Q5JVZ0, Q70LR8, Q70LS7, Q70LS8, Q70LS9, Q70LT7, Q70LT8, Q70LT9, Q70LU0, Q70LU1, Q96RV4, Q96RW0, Q9BZA6, Q9H4E0, Q9P2M0, Q9P2X5 | Gene names | PCDH11X, PCDH11, PCDHX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 X-linked precursor (Protocadherin-11) (Protocadherin on the X chromosome) (PCDH-X) (Protocadherin-S). | |||||
|
SEMG1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.017798 (rank : 42) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P04279, Q6X4I9, Q6Y809, Q6Y822, Q6Y823, Q86U64, Q96QM3 | Gene names | SEMG1, SEMG | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-1 precursor (Semenogelin I) (SGI) [Contains: Alpha- inhibin-92; Alpha-inhibin-31; Seminal basic protein]. | |||||
|
WDR60_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.027667 (rank : 38) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WVS4, Q9NW58 | Gene names | WDR60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
ZN638_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.011933 (rank : 50) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
SCML2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.007779 (rank : 61) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQR0, Q86U98, Q8IWD0, Q8NDP2, Q9UGC5 | Gene names | SCML2 | |||
|
Domain Architecture |

|
|||||
| Description | Sex comb on midleg-like protein 2. | |||||
|
ESCO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.013595 (rank : 47) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
FANCM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.010450 (rank : 53) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
KIF11_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.000997 (rank : 78) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P9P6, Q9Z1J0 | Gene names | Kif11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF11 (Kinesin-related motor protein Eg5). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.013116 (rank : 48) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.009185 (rank : 56) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SENP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.006868 (rank : 63) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59110 | Gene names | Senp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 1 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP1). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.008895 (rank : 57) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ASAH1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.011705 (rank : 51) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WV54 | Gene names | Asah1, Asah | |||
|
Domain Architecture |

|
|||||
| Description | Acid ceramidase precursor (EC 3.5.1.23) (Acylsphingosine deacylase) (N-acylsphingosine amidohydrolase) (AC) [Contains: Acid ceramidase subunit alpha; Acid ceramidase subunit beta]. | |||||
|
OSTP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.009791 (rank : 54) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P10923, P19008 | Gene names | Spp1, Eta-1, Op, Spp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Osteopontin precursor (Bone sialoprotein-1) (Secreted phosphoprotein 1) (SPP-1) (Minopontin) (Early T-lymphocyte activation 1 protein) (2AR) (Calcium oxalate crystal growth inhibitor protein). | |||||
|
SEMG2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.010714 (rank : 52) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02383, Q6X2M6 | Gene names | SEMG2 | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-2 precursor (Semenogelin II) (SGII). | |||||
|
SFRS6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.009726 (rank : 55) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.008434 (rank : 59) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
TRI59_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.004971 (rank : 70) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWR1 | Gene names | TRIM59, RNF104, TRIM57, TSBF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 59 (Tumor suppressor TSBF-1) (RING finger protein 104). | |||||
|
ZSCA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | -0.004072 (rank : 82) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z7L9, Q6ZQY9, Q9NWU4 | Gene names | ZSCAN2, ZFP29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 2 (Zinc finger protein 29 homolog) (Zfp-29). | |||||
|
ZSCA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | -0.004051 (rank : 81) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 767 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07230 | Gene names | Zscan2, Zfp-29, Zfp29 | |||
|
Domain Architecture |
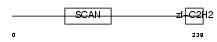
|
|||||
| Description | Zinc finger and SCAN domain-containing protein 2 (Zinc finger protein 29) (Zfp-29). | |||||
|
AFF3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.005605 (rank : 66) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
FIP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.014089 (rank : 46) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6UN15, Q499Y4, Q49AU3, Q7Z608, Q8WVN3, Q96F80, Q9H077 | Gene names | FIP1L1, FIP1, RHE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1) (Factor interacting with PAP) (hFip1) (Rearranged in hypereosinophilia). | |||||
|
FOXP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.001234 (rank : 77) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DBY0, Q80V92, Q8CG10, Q8CIS1 | Gene names | Foxp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein P4 (Fork head-related protein like A) (mFKHLA). | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.005149 (rank : 69) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
SFR14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.005222 (rank : 68) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | -0.006024 (rank : 84) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TRI59_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.004640 (rank : 72) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q922Y2, Q9CSP2, Q9CUD5, Q9D740 | Gene names | Trim59, Mrf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 59 (RING finger protein 1). | |||||
|
TS1R1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.042548 (rank : 36) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99PG6, Q6NS58, Q923J9, Q925I5, Q99PG5 | Gene names | Tas1r1, Gpr70, T1r1, Tr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.003452 (rank : 73) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.007860 (rank : 60) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
APC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.008730 (rank : 58) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
CA026_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.005633 (rank : 65) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBQ9, Q6PAN2, Q7TMQ6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26 homolog. | |||||
|
EXOSX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.005376 (rank : 67) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01780, Q15158 | Gene names | EXOSC10, PMSCL, PMSCL2 | |||
|
Domain Architecture |

|
|||||
| Description | Exosome component 10 (Polymyositis/scleroderma autoantigen 2) (Autoantigen PM/Scl 2) (Polymyositis/scleroderma autoantigen 100 kDa) (PM/Scl-100) (P100 polymyositis-scleroderma overlap syndrome- associated autoantigen). | |||||
|
FLRT3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | -0.001164 (rank : 80) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NZU0, Q96K39, Q96K42, Q96KB1, Q9P259 | Gene names | FLRT3, KIAA1469 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat transmembrane protein FLRT3 precursor (Fibronectin-like domain-containing leucine-rich transmembrane protein 3). | |||||
|
KLOTB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.003026 (rank : 74) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99N32, Q920J2, Q99N31 | Gene names | Klb, Betakl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta klotho (BetaKlotho) (Klotho beta-like protein). | |||||
|
PC11Y_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.000583 (rank : 79) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZA8, Q70LR6, Q70LR8, Q70LS0, Q70LS1, Q70LS2, Q70LS3, Q70LS4, Q70LS5, Q8WY34, Q9BZA9, Q9H4E1 | Gene names | PCDH11Y, PCDH11, PCDH22, PCDHY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 Y-linked precursor (Protocadherin-11) (Protocadherin- 22) (Protocadherin on the Y chromosome) (PCDH-Y) (Protocadherin prostate cancer) (Protocadherin-PC). | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.004727 (rank : 71) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
ZCHC6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.007552 (rank : 62) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5BLK4, Q8CIH3 | Gene names | Zcchc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
ZN595_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | -0.004335 (rank : 83) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 798 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IYB9 | Gene names | ZNF595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 595. | |||||
|
NMDE1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P35436 | Gene names | Grin2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A). | |||||
|
NMDE1_HUMAN
|
||||||
| NC score | 0.998225 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q12879, O00669 | Gene names | GRIN2A | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A) (hNR2A). | |||||
|
NMDE2_HUMAN
|
||||||
| NC score | 0.992656 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13224, Q12919, Q13220, Q13225, Q9UM56 | Gene names | GRIN2B | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B) (N-methyl-D-aspartate receptor subunit 3) (NR3) (hNR3). | |||||
|
NMDE2_MOUSE
|
||||||
| NC score | 0.992493 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q01097, Q9DCB2 | Gene names | Grin2b | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B). | |||||
|
NMDE3_MOUSE
|
||||||
| NC score | 0.987585 (rank : 5) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q01098 | Gene names | Grin2c | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE3_HUMAN
|
||||||
| NC score | 0.987121 (rank : 6) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE4_HUMAN
|
||||||
| NC score | 0.984920 (rank : 7) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
|
NMDE4_MOUSE
|
||||||
| NC score | 0.982499 (rank : 8) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q03391 | Gene names | Grin2d | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D). | |||||
|
NMD3A_HUMAN
|
||||||
| NC score | 0.933847 (rank : 9) | θ value | 2.47191e-82 (rank : 9) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TCU5, Q8TF29, Q8WXI6 | Gene names | GRIN3A, KIAA1973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3A precursor (N-methyl-D-aspartate receptor subtype NR3A) (NMDAR-L). | |||||
|
NMD3B_MOUSE
|
||||||
| NC score | 0.925136 (rank : 10) | θ value | 7.45998e-71 (rank : 12) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91ZU9, Q8VHV0 | Gene names | Grin3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subunit NR3B) (NR3B) (NMDAR3B) (NMDA receptor NR4) (Nr4). | |||||
|
NMD3B_HUMAN
|
||||||
| NC score | 0.923653 (rank : 11) | θ value | 5.00389e-67 (rank : 13) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60391, Q5EAK7, Q7RTW9 | Gene names | GRIN3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subtype NR3B) (NR3B) (NMDAR3B). | |||||
|
NMDZ1_HUMAN
|
||||||
| NC score | 0.908337 (rank : 12) | θ value | 2.0926e-81 (rank : 10) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q05586, P35437, Q12867, Q12868 | Gene names | GRIN1, NMDAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMDZ1_MOUSE
|
||||||
| NC score | 0.907859 (rank : 13) | θ value | 4.66183e-81 (rank : 11) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35438, Q8CFS4 | Gene names | Grin1, Glurz1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
GRID2_MOUSE
|
||||||
| NC score | 0.843840 (rank : 14) | θ value | 2.41655e-45 (rank : 17) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61625 | Gene names | Grid2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRID2_HUMAN
|
||||||
| NC score | 0.842495 (rank : 15) | θ value | 5.95217e-44 (rank : 24) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43424 | Gene names | GRID2, GLURD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRID1_MOUSE
|
||||||
| NC score | 0.839376 (rank : 16) | θ value | 1.79215e-40 (rank : 31) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61627 | Gene names | Grid1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRIK3_HUMAN
|
||||||
| NC score | 0.838284 (rank : 17) | θ value | 7.51208e-47 (rank : 15) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13003, Q13004, Q16136 | Gene names | GRIK3, GLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 3 precursor (Glutamate receptor 7) (GluR-7) (GluR7) (Excitatory amino acid receptor 5) (EAA5). | |||||
|
GRID1_HUMAN
|
||||||
| NC score | 0.837835 (rank : 18) | θ value | 8.89434e-40 (rank : 32) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULK0, Q8IXT3 | Gene names | GRID1, KIAA1220 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRIK1_HUMAN
|
||||||
| NC score | 0.837510 (rank : 19) | θ value | 3.72821e-46 (rank : 16) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P39086, Q13001, Q86SU9 | Gene names | GRIK1, GLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5) (Excitatory amino acid receptor 3) (EAA3). | |||||
|
GRIK4_HUMAN
|
||||||
| NC score | 0.836907 (rank : 20) | θ value | 1.32601e-43 (rank : 25) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16099 | Gene names | GRIK4, GRIK | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor (Glutamate receptor KA-1) (KA1) (Excitatory amino acid receptor 1) (EAA1). | |||||
|
GRIK4_MOUSE
|
||||||
| NC score | 0.836663 (rank : 21) | θ value | 1.32601e-43 (rank : 26) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BMF5 | Gene names | Grik4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor. | |||||
|
GRIK2_HUMAN
|
||||||
| NC score | 0.836174 (rank : 22) | θ value | 1.51363e-47 (rank : 14) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13002 | Gene names | GRIK2, GLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 2 precursor (Glutamate receptor 6) (GluR-6) (GluR6) (Excitatory amino acid receptor 4) (EAA4). | |||||
|
GRIK5_HUMAN
|
||||||
| NC score | 0.836080 (rank : 23) | θ value | 7.03111e-45 (rank : 19) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16478 | Gene names | GRIK5, GRIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Excitatory amino acid receptor 2) (EAA2). | |||||
|
GRIK5_MOUSE
|
||||||
| NC score | 0.835594 (rank : 24) | θ value | 1.19932e-44 (rank : 20) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61626 | Gene names | Grik5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Glutamate receptor gamma-2) (GluR gamma-2). | |||||
|
GRIA1_MOUSE
|
||||||
| NC score | 0.830701 (rank : 25) | θ value | 7.03111e-45 (rank : 18) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23818 | Gene names | Gria1, Glur1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIA1_HUMAN
|
||||||
| NC score | 0.829335 (rank : 26) | θ value | 2.04573e-44 (rank : 21) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42261 | Gene names | GRIA1, GLUH1, GLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIA4_HUMAN
|
||||||
| NC score | 0.827156 (rank : 27) | θ value | 8.59492e-43 (rank : 27) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48058 | Gene names | GRIA4, GLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIA3_HUMAN
|
||||||
| NC score | 0.825308 (rank : 28) | θ value | 2.67181e-44 (rank : 22) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42263, Q4VXD5, Q4VXD6, Q9HDA0, Q9HDA1, Q9HDA2 | Gene names | GRIA3, GLUR3, GLURC | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRIA2_HUMAN
|
||||||
| NC score | 0.825307 (rank : 29) | θ value | 2.50074e-42 (rank : 28) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42262, Q96FP6 | Gene names | GRIA2, GLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIA3_MOUSE
|
||||||
| NC score | 0.825288 (rank : 30) | θ value | 4.55743e-44 (rank : 23) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2W9, Q5DTJ0 | Gene names | Gria3, Glur3, Kiaa4184 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRIA4_MOUSE
|
||||||
| NC score | 0.825008 (rank : 31) | θ value | 9.50281e-42 (rank : 30) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2W8 | Gene names | Gria4, Glur4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIA2_MOUSE
|
||||||
| NC score | 0.824644 (rank : 32) | θ value | 4.2656e-42 (rank : 29) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23819, Q61604, Q61605, Q8BG69, Q8BXU3, Q9D6D3 | Gene names | Gria2, Glur2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIK1_MOUSE
|
||||||
| NC score | 0.818341 (rank : 33) | θ value | 1.37539e-32 (rank : 33) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q60934 | Gene names | Grik1, Glur5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5). | |||||
|
ERB1_HUMAN
|
||||||
| NC score | 0.139982 (rank : 34) | θ value | 0.0563607 (rank : 35) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60509 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-R(b)_3p24.3 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-R(b) Env protein) [Includes: Surface protein domain (SU); Transmembrane protein domain (TM)]. | |||||
|
DLEC1_MOUSE
|
||||||
| NC score | 0.054166 (rank : 35) | θ value | 0.0252991 (rank : 34) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BLA1 | Gene names | Dlec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in lung and esophageal cancer protein 1 homolog. | |||||
|
TS1R1_MOUSE
|
||||||
| NC score | 0.042548 (rank : 36) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99PG6, Q6NS58, Q923J9, Q925I5, Q99PG5 | Gene names | Tas1r1, Gpr70, T1r1, Tr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70). | |||||
|
CIR_HUMAN
|
||||||
| NC score | 0.029361 (rank : 37) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
WDR60_HUMAN
|
||||||
| NC score | 0.027667 (rank : 38) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WVS4, Q9NW58 | Gene names | WDR60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.025394 (rank : 39) | θ value | 0.125558 (rank : 37) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.021089 (rank : 40) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
S19A3_MOUSE
|
||||||
| NC score | 0.020137 (rank : 41) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99PL8, Q32MF5 | Gene names | Slc19a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thiamine transporter 2 (ThTr-2) (ThTr2) (Solute carrier family 19 member 3). | |||||
|
SEMG1_HUMAN
|
||||||
| NC score | 0.017798 (rank : 42) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P04279, Q6X4I9, Q6Y809, Q6Y822, Q6Y823, Q86U64, Q96QM3 | Gene names | SEMG1, SEMG | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-1 precursor (Semenogelin I) (SGI) [Contains: Alpha- inhibin-92; Alpha-inhibin-31; Seminal basic protein]. | |||||
|
CKAP2_HUMAN
|
||||||
| NC score | 0.017354 (rank : 43) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WWK9, Q3KRA5, Q5VXB4, Q8IWV5, Q8IWV6, Q96FH9, Q9H012, Q9H0D0, Q9H988, Q9HC49, Q9NVG4 | Gene names | CKAP2, LB1, TMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 2 (Tumor-associated microtubule- associated protein) (CTCL tumor antigen se20-10). | |||||
|
ERCC6_HUMAN
|
||||||
| NC score | 0.016183 (rank : 44) | θ value | 0.279714 (rank : 38) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03468 | Gene names | ERCC6, CSB | |||
|
Domain Architecture |

|
|||||
| Description | DNA excision repair protein ERCC-6 (EC 3.6.1.-) (ATP-dependent helicase ERCC6) (Cockayne syndrome protein CSB). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.015338 (rank : 45) | θ value | 0.0961366 (rank : 36) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
FIP1_HUMAN
|
||||||
| NC score | 0.014089 (rank : 46) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6UN15, Q499Y4, Q49AU3, Q7Z608, Q8WVN3, Q96F80, Q9H077 | Gene names | FIP1L1, FIP1, RHE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1) (Factor interacting with PAP) (hFip1) (Rearranged in hypereosinophilia). | |||||
|
ESCO1_HUMAN
|
||||||
| NC score | 0.013595 (rank : 47) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.013116 (rank : 48) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.011934 (rank : 49) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
ZN638_MOUSE
|
||||||
| NC score | 0.011933 (rank : 50) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
ASAH1_MOUSE
|
||||||
| NC score | 0.011705 (rank : 51) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WV54 | Gene names | Asah1, Asah | |||
|
Domain Architecture |

|
|||||
| Description | Acid ceramidase precursor (EC 3.5.1.23) (Acylsphingosine deacylase) (N-acylsphingosine amidohydrolase) (AC) [Contains: Acid ceramidase subunit alpha; Acid ceramidase subunit beta]. | |||||
|
SEMG2_HUMAN
|
||||||
| NC score | 0.010714 (rank : 52) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02383, Q6X2M6 | Gene names | SEMG2 | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-2 precursor (Semenogelin II) (SGII). | |||||
|
FANCM_MOUSE
|
||||||
| NC score | 0.010450 (rank : 53) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
OSTP_MOUSE
|
||||||
| NC score | 0.009791 (rank : 54) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P10923, P19008 | Gene names | Spp1, Eta-1, Op, Spp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Osteopontin precursor (Bone sialoprotein-1) (Secreted phosphoprotein 1) (SPP-1) (Minopontin) (Early T-lymphocyte activation 1 protein) (2AR) (Calcium oxalate crystal growth inhibitor protein). | |||||
|
SFRS6_HUMAN
|
||||||
| NC score | 0.009726 (rank : 55) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.009185 (rank : 56) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.008895 (rank : 57) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.008730 (rank : 58) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.008434 (rank : 59) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.007860 (rank : 60) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
SCML2_HUMAN
|
||||||
| NC score | 0.007779 (rank : 61) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQR0, Q86U98, Q8IWD0, Q8NDP2, Q9UGC5 | Gene names | SCML2 | |||
|
Domain Architecture |

|
|||||
| Description | Sex comb on midleg-like protein 2. | |||||
|
ZCHC6_MOUSE
|
||||||
| NC score | 0.007552 (rank : 62) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5BLK4, Q8CIH3 | Gene names | Zcchc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
SENP1_MOUSE
|
||||||
| NC score | 0.006868 (rank : 63) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59110 | Gene names | Senp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 1 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP1). | |||||
|
GALT5_HUMAN
|
||||||
| NC score | 0.006660 (rank : 64) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z7M9, Q9UGK7, Q9UHL6 | Gene names | GALNT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 5) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 5) (Polypeptide GalNAc transferase 5) (GalNAc-T5) (pp-GaNTase 5). | |||||
|
CA026_MOUSE
|
||||||
| NC score | 0.005633 (rank : 65) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBQ9, Q6PAN2, Q7TMQ6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26 homolog. | |||||
|
AFF3_HUMAN
|
||||||
| NC score | 0.005605 (rank : 66) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
EXOSX_HUMAN
|
||||||
| NC score | 0.005376 (rank : 67) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01780, Q15158 | Gene names | EXOSC10, PMSCL, PMSCL2 | |||
|
Domain Architecture |

|
|||||
| Description | Exosome component 10 (Polymyositis/scleroderma autoantigen 2) (Autoantigen PM/Scl 2) (Polymyositis/scleroderma autoantigen 100 kDa) (PM/Scl-100) (P100 polymyositis-scleroderma overlap syndrome- associated autoantigen). | |||||
|
SFR14_MOUSE
|
||||||
| NC score | 0.005222 (rank : 68) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.005149 (rank : 69) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
TRI59_HUMAN
|
||||||
| NC score | 0.004971 (rank : 70) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWR1 | Gene names | TRIM59, RNF104, TRIM57, TSBF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 59 (Tumor suppressor TSBF-1) (RING finger protein 104). | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.004727 (rank : 71) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
TRI59_MOUSE
|
||||||
| NC score | 0.004640 (rank : 72) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q922Y2, Q9CSP2, Q9CUD5, Q9D740 | Gene names | Trim59, Mrf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 59 (RING finger protein 1). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.003452 (rank : 73) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
KLOTB_MOUSE
|
||||||
| NC score | 0.003026 (rank : 74) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99N32, Q920J2, Q99N31 | Gene names | Klb, Betakl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta klotho (BetaKlotho) (Klotho beta-like protein). | |||||
|
PC11X_HUMAN
|
||||||
| NC score | 0.001644 (rank : 75) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZA7, Q2TJH0, Q2TJH1, Q2TJH3, Q5JVZ0, Q70LR8, Q70LS7, Q70LS8, Q70LS9, Q70LT7, Q70LT8, Q70LT9, Q70LU0, Q70LU1, Q96RV4, Q96RW0, Q9BZA6, Q9H4E0, Q9P2M0, Q9P2X5 | Gene names | PCDH11X, PCDH11, PCDHX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 X-linked precursor (Protocadherin-11) (Protocadherin on the X chromosome) (PCDH-X) (Protocadherin-S). | |||||
|
WNK3_HUMAN
|
||||||
| NC score | 0.001615 (rank : 76) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYP7, Q9HCK6 | Gene names | WNK3, KIAA1566, PRKWNK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK3 (EC 2.7.11.1) (Protein kinase with no lysine 3) (Protein kinase, lysine-deficient 3). | |||||
|
FOXP4_MOUSE
|
||||||
| NC score | 0.001234 (rank : 77) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DBY0, Q80V92, Q8CG10, Q8CIS1 | Gene names | Foxp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein P4 (Fork head-related protein like A) (mFKHLA). | |||||
|
KIF11_MOUSE
|
||||||
| NC score | 0.000997 (rank : 78) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P9P6, Q9Z1J0 | Gene names | Kif11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF11 (Kinesin-related motor protein Eg5). | |||||
|
PC11Y_HUMAN
|
||||||
| NC score | 0.000583 (rank : 79) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZA8, Q70LR6, Q70LR8, Q70LS0, Q70LS1, Q70LS2, Q70LS3, Q70LS4, Q70LS5, Q8WY34, Q9BZA9, Q9H4E1 | Gene names | PCDH11Y, PCDH11, PCDH22, PCDHY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 Y-linked precursor (Protocadherin-11) (Protocadherin- 22) (Protocadherin on the Y chromosome) (PCDH-Y) (Protocadherin prostate cancer) (Protocadherin-PC). | |||||
|
FLRT3_HUMAN
|
||||||
| NC score | -0.001164 (rank : 80) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NZU0, Q96K39, Q96K42, Q96KB1, Q9P259 | Gene names | FLRT3, KIAA1469 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat transmembrane protein FLRT3 precursor (Fibronectin-like domain-containing leucine-rich transmembrane protein 3). | |||||
|
ZSCA2_MOUSE
|
||||||
| NC score | -0.004051 (rank : 81) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 767 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07230 | Gene names | Zscan2, Zfp-29, Zfp29 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and SCAN domain-containing protein 2 (Zinc finger protein 29) (Zfp-29). | |||||
|
ZSCA2_HUMAN
|
||||||
| NC score | -0.004072 (rank : 82) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z7L9, Q6ZQY9, Q9NWU4 | Gene names | ZSCAN2, ZFP29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 2 (Zinc finger protein 29 homolog) (Zfp-29). | |||||
|
ZN595_HUMAN
|
||||||
| NC score | -0.004335 (rank : 83) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 798 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IYB9 | Gene names | ZNF595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 595. | |||||
|
TITIN_HUMAN
|
||||||
| NC score | -0.006024 (rank : 84) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||