Please be patient as the page loads
|
LAP2A_MOUSE
|
||||||
| SwissProt Accessions | Q61033, Q61028 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms alpha/zeta (Thymopoietin isoforms alpha/zeta) (TP alpha/zeta). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LAP2A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.987922 (rank : 2) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P42166, P08918, P08919, Q14860, Q16295 | Gene names | TMPO, LAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoform alpha (Thymopoietin isoform alpha) (TP alpha) (Thymopoietin-related peptide isoform alpha) (TPRP isoform alpha) [Contains: Thymopoietin (TP) (Splenin); Thymopentin (TP5)]. | |||||
|
LAP2A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61033, Q61028 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms alpha/zeta (Thymopoietin isoforms alpha/zeta) (TP alpha/zeta). | |||||
|
LAP2B_MOUSE
|
||||||
| θ value | 6.95003e-85 (rank : 3) | NC score | 0.925510 (rank : 3) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61029, Q61030, Q61031, Q61032 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms beta/delta/epsilon/gamma (Thymopoietin isoforms beta/delta/epsilon/gamma) (TP beta/delta/epsilon/gamma). | |||||
|
LAP2B_HUMAN
|
||||||
| θ value | 8.22888e-78 (rank : 4) | NC score | 0.911662 (rank : 4) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P42167, Q14861 | Gene names | TMPO, LAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2, isoforms beta/gamma (Thymopoietin, isoforms beta/gamma) (TP beta/gamma) (Thymopoietin-related peptide isoforms beta/gamma) (TPRP isoforms beta/gamma) [Contains: Thymopoietin (TP) (Splenin); Thymopentin (TP5)]. | |||||
|
EMD_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 5) | NC score | 0.291719 (rank : 6) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P50402 | Gene names | EMD, EDMD, STA | |||
|
Domain Architecture |

|
|||||
| Description | Emerin. | |||||
|
EMD_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 6) | NC score | 0.298350 (rank : 5) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08579 | Gene names | Emd, Sta | |||
|
Domain Architecture |

|
|||||
| Description | Emerin. | |||||
|
MAN1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 7) | NC score | 0.181885 (rank : 7) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2U8, Q9NT47, Q9NYA5 | Gene names | LEMD3, MAN1 | |||
|
Domain Architecture |
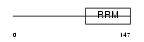
|
|||||
| Description | Inner nuclear membrane protein Man1 (LEM domain-containing protein 3). | |||||
|
FNBP2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 8) | NC score | 0.026086 (rank : 12) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
LARP1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 9) | NC score | 0.103346 (rank : 9) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
CCNI_HUMAN
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.110410 (rank : 8) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14094 | Gene names | CCNI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
M3K14_MOUSE
|
||||||
| θ value | 0.365318 (rank : 11) | NC score | 0.016276 (rank : 17) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WUL6 | Gene names | Map3k14, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 14 (EC 2.7.11.25) (NF- kappa beta-inducing kinase) (Serine/threonine-protein kinase NIK). | |||||
|
SON_MOUSE
|
||||||
| θ value | 0.47712 (rank : 12) | NC score | 0.025164 (rank : 14) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
CN104_MOUSE
|
||||||
| θ value | 0.813845 (rank : 13) | NC score | 0.045630 (rank : 11) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BPI1, Q9CZC7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf104 homolog. | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 14) | NC score | 0.006184 (rank : 33) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
AT2C2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.024909 (rank : 15) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75185 | Gene names | KIAA0703 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable calcium-transporting ATPase KIAA0703 (EC 3.6.3.8). | |||||
|
HIPK1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.009961 (rank : 26) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88904, Q80TV5, Q9QUQ8, Q9QZR3, Q9WVN7 | Gene names | Hipk1, Kiaa0630, Myak, Nbak2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeodomain-interacting protein kinase 1 (EC 2.7.11.1) (Protein kinase Myak) (Nuclear body-associated kinase 2). | |||||
|
PASK_MOUSE
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.007399 (rank : 30) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CEE6, Q91YD8 | Gene names | Pask, Kiaa0135 | |||
|
Domain Architecture |

|
|||||
| Description | PAS domain-containing serine/threonine-protein kinase (EC 2.7.11.1) (PAS-kinase) (PASKIN). | |||||
|
XRN1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.025812 (rank : 13) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97789, O35651, P97790, Q3TE44, Q3TRE9, Q3UQL5 | Gene names | Xrn1, Dhm2, Exo, Sep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-3' exoribonuclease 1 (EC 3.1.11.-) (mXRN1) (Strand-exchange protein 1 homolog) (Protein Dhm2). | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.005740 (rank : 34) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
ZFP37_HUMAN
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.005419 (rank : 37) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 797 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6Q3 | Gene names | ZFP37 | |||
|
Domain Architecture |
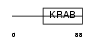
|
|||||
| Description | Zinc finger protein 37 homolog (Zfp-37). | |||||
|
M3K14_HUMAN
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.011424 (rank : 21) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99558, Q8IYN1 | Gene names | MAP3K14, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 14 (EC 2.7.11.25) (NF- kappa beta-inducing kinase) (Serine/threonine-protein kinase NIK) (HsNIK). | |||||
|
FOXN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.010445 (rank : 25) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
HIPK1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.008782 (rank : 28) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86Z02, O75125, Q8IYD7, Q8NDN5, Q8NEB6, Q8TBZ1 | Gene names | HIPK1, KIAA0630 | |||
|
Domain Architecture |

|
|||||
| Description | Homeodomain-interacting protein kinase 1 (EC 2.7.11.1). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.010695 (rank : 24) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
RIMS2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.010981 (rank : 23) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
WBS14_MOUSE
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.019539 (rank : 16) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
CCNI_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.063481 (rank : 10) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2V9 | Gene names | Ccni | |||
|
Domain Architecture |
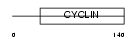
|
|||||
| Description | Cyclin-I. | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.004814 (rank : 38) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
PITX2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.005696 (rank : 35) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99697, O60578, O60579, O60580, Q9BY17 | Gene names | PITX2, ARP1, RGS, RIEG, RIEG1 | |||
|
Domain Architecture |
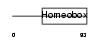
|
|||||
| Description | Pituitary homeobox 2 (RIEG bicoid-related homeobox transcription factor) (Solurshin) (ALL1-responsive protein ARP1). | |||||
|
PITX2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.005547 (rank : 36) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97474, O08646, O70336, P97933, Q9JLA0, Q9QXB8, Q9R1V9, Q9Z141 | Gene names | Pitx2, Arp1, Brx1, Otlx2, Ptx2, Rgs | |||
|
Domain Architecture |
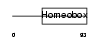
|
|||||
| Description | Pituitary homeobox 2 (Orthodenticle-like homeobox 2) (Solurshin) (ALL1-responsive protein ARP1) (BRX1 homeoprotein) (Paired-like homeodomain transcription factor Munc 30). | |||||
|
SNIP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.013894 (rank : 18) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9C0H9, Q8N4W8 | Gene names | P140, KIAA1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
VAV3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.006755 (rank : 31) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKW4, O95230, Q9Y5X8 | Gene names | VAV3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
ZC3H3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.013677 (rank : 19) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
AKAP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.011391 (rank : 22) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
ASXL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.011758 (rank : 20) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59598 | Gene names | Asxl1, Kiaa0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
DDX24_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.006405 (rank : 32) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.009780 (rank : 27) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.007919 (rank : 29) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
LAP2A_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61033, Q61028 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms alpha/zeta (Thymopoietin isoforms alpha/zeta) (TP alpha/zeta). | |||||
|
LAP2A_HUMAN
|
||||||
| NC score | 0.987922 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P42166, P08918, P08919, Q14860, Q16295 | Gene names | TMPO, LAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoform alpha (Thymopoietin isoform alpha) (TP alpha) (Thymopoietin-related peptide isoform alpha) (TPRP isoform alpha) [Contains: Thymopoietin (TP) (Splenin); Thymopentin (TP5)]. | |||||
|
LAP2B_MOUSE
|
||||||
| NC score | 0.925510 (rank : 3) | θ value | 6.95003e-85 (rank : 3) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61029, Q61030, Q61031, Q61032 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms beta/delta/epsilon/gamma (Thymopoietin isoforms beta/delta/epsilon/gamma) (TP beta/delta/epsilon/gamma). | |||||
|
LAP2B_HUMAN
|
||||||
| NC score | 0.911662 (rank : 4) | θ value | 8.22888e-78 (rank : 4) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P42167, Q14861 | Gene names | TMPO, LAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2, isoforms beta/gamma (Thymopoietin, isoforms beta/gamma) (TP beta/gamma) (Thymopoietin-related peptide isoforms beta/gamma) (TPRP isoforms beta/gamma) [Contains: Thymopoietin (TP) (Splenin); Thymopentin (TP5)]. | |||||
|
EMD_MOUSE
|
||||||
| NC score | 0.298350 (rank : 5) | θ value | 0.00134147 (rank : 6) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08579 | Gene names | Emd, Sta | |||
|
Domain Architecture |

|
|||||
| Description | Emerin. | |||||
|
EMD_HUMAN
|
||||||
| NC score | 0.291719 (rank : 6) | θ value | 0.00134147 (rank : 5) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P50402 | Gene names | EMD, EDMD, STA | |||
|
Domain Architecture |

|
|||||
| Description | Emerin. | |||||
|
MAN1_HUMAN
|
||||||
| NC score | 0.181885 (rank : 7) | θ value | 0.0113563 (rank : 7) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2U8, Q9NT47, Q9NYA5 | Gene names | LEMD3, MAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Inner nuclear membrane protein Man1 (LEM domain-containing protein 3). | |||||
|
CCNI_HUMAN
|
||||||
| NC score | 0.110410 (rank : 8) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14094 | Gene names | CCNI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
LARP1_HUMAN
|
||||||
| NC score | 0.103346 (rank : 9) | θ value | 0.0961366 (rank : 9) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
CCNI_MOUSE
|
||||||
| NC score | 0.063481 (rank : 10) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2V9 | Gene names | Ccni | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
CN104_MOUSE
|
||||||
| NC score | 0.045630 (rank : 11) | θ value | 0.813845 (rank : 13) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BPI1, Q9CZC7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf104 homolog. | |||||
|
FNBP2_HUMAN
|
||||||
| NC score | 0.026086 (rank : 12) | θ value | 0.0961366 (rank : 8) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
XRN1_MOUSE
|
||||||
| NC score | 0.025812 (rank : 13) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97789, O35651, P97790, Q3TE44, Q3TRE9, Q3UQL5 | Gene names | Xrn1, Dhm2, Exo, Sep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-3' exoribonuclease 1 (EC 3.1.11.-) (mXRN1) (Strand-exchange protein 1 homolog) (Protein Dhm2). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.025164 (rank : 14) | θ value | 0.47712 (rank : 12) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
AT2C2_HUMAN
|
||||||
| NC score | 0.024909 (rank : 15) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75185 | Gene names | KIAA0703 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable calcium-transporting ATPase KIAA0703 (EC 3.6.3.8). | |||||
|
WBS14_MOUSE
|
||||||
| NC score | 0.019539 (rank : 16) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
M3K14_MOUSE
|
||||||
| NC score | 0.016276 (rank : 17) | θ value | 0.365318 (rank : 11) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WUL6 | Gene names | Map3k14, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 14 (EC 2.7.11.25) (NF- kappa beta-inducing kinase) (Serine/threonine-protein kinase NIK). | |||||
|
SNIP_HUMAN
|
||||||
| NC score | 0.013894 (rank : 18) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9C0H9, Q8N4W8 | Gene names | P140, KIAA1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
ZC3H3_MOUSE
|
||||||
| NC score | 0.013677 (rank : 19) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ASXL1_MOUSE
|
||||||
| NC score | 0.011758 (rank : 20) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59598 | Gene names | Asxl1, Kiaa0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
M3K14_HUMAN
|
||||||
| NC score | 0.011424 (rank : 21) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99558, Q8IYN1 | Gene names | MAP3K14, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 14 (EC 2.7.11.25) (NF- kappa beta-inducing kinase) (Serine/threonine-protein kinase NIK) (HsNIK). | |||||
|
AKAP1_HUMAN
|
||||||
| NC score | 0.011391 (rank : 22) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
RIMS2_HUMAN
|
||||||
| NC score | 0.010981 (rank : 23) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.010695 (rank : 24) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
FOXN1_MOUSE
|
||||||
| NC score | 0.010445 (rank : 25) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
HIPK1_MOUSE
|
||||||
| NC score | 0.009961 (rank : 26) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88904, Q80TV5, Q9QUQ8, Q9QZR3, Q9WVN7 | Gene names | Hipk1, Kiaa0630, Myak, Nbak2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeodomain-interacting protein kinase 1 (EC 2.7.11.1) (Protein kinase Myak) (Nuclear body-associated kinase 2). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.009780 (rank : 27) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
HIPK1_HUMAN
|
||||||
| NC score | 0.008782 (rank : 28) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86Z02, O75125, Q8IYD7, Q8NDN5, Q8NEB6, Q8TBZ1 | Gene names | HIPK1, KIAA0630 | |||
|
Domain Architecture |

|
|||||
| Description | Homeodomain-interacting protein kinase 1 (EC 2.7.11.1). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.007919 (rank : 29) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PASK_MOUSE
|
||||||
| NC score | 0.007399 (rank : 30) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CEE6, Q91YD8 | Gene names | Pask, Kiaa0135 | |||
|
Domain Architecture |

|
|||||
| Description | PAS domain-containing serine/threonine-protein kinase (EC 2.7.11.1) (PAS-kinase) (PASKIN). | |||||
|
VAV3_HUMAN
|
||||||
| NC score | 0.006755 (rank : 31) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKW4, O95230, Q9Y5X8 | Gene names | VAV3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
DDX24_MOUSE
|
||||||
| NC score | 0.006405 (rank : 32) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.006184 (rank : 33) | θ value | 1.06291 (rank : 14) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | 0.005740 (rank : 34) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
PITX2_HUMAN
|
||||||
| NC score | 0.005696 (rank : 35) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99697, O60578, O60579, O60580, Q9BY17 | Gene names | PITX2, ARP1, RGS, RIEG, RIEG1 | |||
|
Domain Architecture |

|
|||||
| Description | Pituitary homeobox 2 (RIEG bicoid-related homeobox transcription factor) (Solurshin) (ALL1-responsive protein ARP1). | |||||
|
PITX2_MOUSE
|
||||||
| NC score | 0.005547 (rank : 36) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97474, O08646, O70336, P97933, Q9JLA0, Q9QXB8, Q9R1V9, Q9Z141 | Gene names | Pitx2, Arp1, Brx1, Otlx2, Ptx2, Rgs | |||
|
Domain Architecture |

|
|||||
| Description | Pituitary homeobox 2 (Orthodenticle-like homeobox 2) (Solurshin) (ALL1-responsive protein ARP1) (BRX1 homeoprotein) (Paired-like homeodomain transcription factor Munc 30). | |||||
|
ZFP37_HUMAN
|
||||||
| NC score | 0.005419 (rank : 37) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 797 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6Q3 | Gene names | ZFP37 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 37 homolog (Zfp-37). | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.004814 (rank : 38) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||