Please be patient as the page loads
|
AKAP1_HUMAN
|
||||||
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AKAP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
AKAP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.938066 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
TDRKH_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 3) | NC score | 0.494539 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y2W6, Q8N582, Q9NYV5 | Gene names | TDRKH, TDRD2 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
TDRKH_MOUSE
|
||||||
| θ value | 7.34386e-10 (rank : 4) | NC score | 0.491036 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80VL1 | Gene names | Tdrkh | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
TDRD7_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 5) | NC score | 0.423705 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NHU6, Q96JT1, Q9UFF0, Q9Y2M3 | Gene names | TDRD7, PCTAIRE2BP | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 7 (Tudor repeat associator with PCTAIRE 2) (Trap). | |||||
|
TDRD7_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 6) | NC score | 0.420218 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K1H1, Q8R181 | Gene names | Tdrd7, Pctaire2bp | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 7 (Tudor repeat associator with PCTAIRE 2) (Trap). | |||||
|
TDRD1_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 7) | NC score | 0.425161 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BXT4, Q9H7B3 | Gene names | TDRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
TDRD1_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 8) | NC score | 0.405354 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99MV1 | Gene names | Tdrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
TDRD4_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 9) | NC score | 0.338642 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NUY9 | Gene names | TDRD4 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 4. | |||||
|
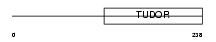
TDRD5_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 10) | NC score | 0.375762 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NAT2, Q5VTV0 | Gene names | TDRD5 | |||
|
Domain Architecture |
|
|||||
| Description | Tudor domain-containing protein 5. | |||||
|
SND1_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 11) | NC score | 0.245638 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7KZF4, Q13122 | Gene names | SND1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Staphylococcal nuclease domain-containing protein 1 (p100 co- activator) (100 kDa coactivator) (EBNA2 coactivator p100). | |||||
|
PQBP1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 12) | NC score | 0.114033 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VJ5, Q80WW2, Q9ER43, Q9QYY2 | Gene names | Pqbp1, Npw38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyglutamine-binding protein 1 (Polyglutamine tract-binding protein 1) (PQBP-1) (38 kDa nuclear protein containing a WW domain) (Npw38). | |||||
|
FUBP3_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 13) | NC score | 0.243925 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96I24, Q92946, Q9BVB6 | Gene names | FUBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 3 (FUSE-binding protein 3). | |||||
|
TDRD6_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 14) | NC score | 0.274732 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P61407 | Gene names | Tdrd6 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 6. | |||||
|
HNRPK_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 15) | NC score | 0.241076 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P61978, Q07244, Q15671, Q60577, Q922Y7, Q96J62 | Gene names | HNRPK, HNRNPK | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K (hnRNP K) (Transformation up-regulated nuclear protein) (TUNP). | |||||
|
HNRPK_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 16) | NC score | 0.241076 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P61979, Q07244, Q15671, Q60577, Q8BGQ8, Q922Y7, Q96J62 | Gene names | Hnrpk, Hnrnpk | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K. | |||||
|
SND1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 17) | NC score | 0.231463 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q78PY7, Q3TT46, Q922L5, Q9R0S1 | Gene names | Snd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Staphylococcal nuclease domain-containing protein 1 (p100 co- activator) (100 kDa coactivator). | |||||
|
PCBP3_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 18) | NC score | 0.219102 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P57721, Q8N9K6, Q96EP6 | Gene names | PCBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 19) | NC score | 0.219176 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P57722, Q8BSB0, Q8C544 | Gene names | Pcbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 20) | NC score | 0.039792 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
PCBP1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 21) | NC score | 0.224697 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15365, Q13157, Q14975 | Gene names | PCBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) (Nucleic acid- binding protein SUB2.3). | |||||
|
PCBP1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 22) | NC score | 0.224697 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P60335 | Gene names | Pcbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1). | |||||
|
NOVA2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 23) | NC score | 0.272156 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UNW9, O43267, Q9UEA1 | Gene names | NOVA2, ANOVA, NOVA3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-2 (Neuro-oncological ventral antigen 2) (Astrocytic NOVA1-like RNA-binding protein). | |||||
|
FUBP2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 24) | NC score | 0.220530 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92945, O00301, Q9UNT5, Q9UQH5 | Gene names | KHSRP, FUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75). | |||||
|
FUBP1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 25) | NC score | 0.219184 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96AE4, Q12828 | Gene names | FUBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP) (DNA helicase V) (HDH V). | |||||
|
FUBP1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 26) | NC score | 0.219596 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91WJ8, Q8C0Y8 | Gene names | Fubp1, D3Ertd330e | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP). | |||||
|
PCBP2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 27) | NC score | 0.216387 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15366, Q6PKG5 | Gene names | PCBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (hnRNP-E2). | |||||
|
PCBP2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 28) | NC score | 0.215765 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61990, Q61383, Q62042 | Gene names | Pcbp2, Cbp, Hnrnpx, Hnrpx | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP). | |||||
|
PDLI7_MOUSE
|
||||||
| θ value | 0.163984 (rank : 29) | NC score | 0.035843 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3TJD7, Q80ZY6, Q810S3, Q8C1S4, Q9CRA1 | Gene names | Pdlim7, Enigma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 30) | NC score | 0.046479 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
NOVA1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.247379 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P51513 | Gene names | NOVA1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-1 (Neuro-oncological ventral antigen 1) (Onconeural ventral antigen 1) (Paraneoplastic Ri antigen) (Ventral neuron-specific protein 1). | |||||
|
MYOZ3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.050411 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R4E4 | Gene names | Myoz3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myozenin-3 (Calsarcin-3) (FATZ-related protein 3). | |||||
|
IF2B2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.183448 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y6M1 | Gene names | IGF2BP2, IMP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2) (Hepatocellular carcinoma autoantigen p62). | |||||
|
PQBP1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.090252 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60828, Q4VY25, Q4VY26, Q4VY27, Q4VY29, Q4VY30, Q4VY34, Q4VY35, Q4VY36, Q4VY37, Q4VY38, Q9GZP2, Q9GZU4, Q9GZZ4 | Gene names | PQBP1, NPW38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyglutamine-binding protein 1 (Polyglutamine tract-binding protein 1) (PQBP-1) (38 kDa nuclear protein containing a WW domain) (Npw38). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.049157 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
MYT1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.030833 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
AN32B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.028200 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.012859 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
HIP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.015409 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
IF2B2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.179759 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5SF07, Q3TCU4, Q7TQF9 | Gene names | Igf2bp2, Imp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2). | |||||
|
PCBP4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.202396 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P57723, Q96AH7 | Gene names | PCBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
PCBP4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.205586 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P57724, Q91VR3 | Gene names | Pcbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
PK3CB_HUMAN
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.036526 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42338 | Gene names | PIK3CB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CB_MOUSE
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.036447 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BTI9 | Gene names | Pik3cb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
CEP41_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.056689 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99NF3, Q8BT53 | Gene names | Tsga14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 41 kDa (Cep41 protein) (Testis-specific gene A14 protein). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.014007 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.017873 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.045268 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
LRRC6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.028911 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86X45, Q13648, Q4G183 | Gene names | LRRC6, LRTP, TSLRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 6 (Leucine-rich testis-specific protein) (Testis-specific leucine-rich repeat protein). | |||||
|
NU153_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.035933 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
SDPR_MOUSE
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.025831 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.040194 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
HSF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.022492 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
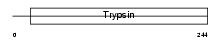
KLK14_HUMAN
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.008842 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P0G3 | Gene names | KLK14, KLKL6 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein-14 precursor (EC 3.4.21.-) (Kallikrein-like protein 6) (KLK-L6). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.048293 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
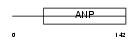
ANF_MOUSE
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.041389 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05125 | Gene names | Nppa, Pnd | |||
|
Domain Architecture |
|
|||||
| Description | Atrial natriuretic factor precursor (ANF) (Atrial natriuretic peptide) (ANP) (Prepronatriodilatin) [Contains: Auriculin-B; Auriculin-A; Atriopeptin I; Atriopeptin II]. | |||||
|
CN037_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.030582 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86TY3, Q6P5Q1, Q86TY1 | Gene names | C14orf37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 precursor. | |||||
|
DDX24_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.011229 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
K0552_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.021583 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60299 | Gene names | KIAA0552 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0552. | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.016337 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
DACH1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.019351 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QYB2, O88716, Q8BPQ0, Q8C8D7, Q9QY47, Q9R218, Q9Z0Y5 | Gene names | Dach1, Dach | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
GOGA5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.011614 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.023209 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
VWF_MOUSE
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.010615 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
FMR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.069857 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q06787, Q16578, Q99054 | Gene names | FMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation 1 protein (Protein FMR-1) (FMRP). | |||||
|
FMR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.069316 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35922 | Gene names | Fmr1, Fmr-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation protein 1 homolog (Protein FMR-1) (FMRP) (mFmr1p). | |||||
|
FRM4A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.016730 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2Q2 | Gene names | FRMD4A, FRMD4, KIAA1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FRM4A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.017180 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BIE6, Q6PDJ4, Q8CHA6 | Gene names | Frmd4a, Frmd4, Kiaa1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FXR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.079409 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P51114, Q7Z450, Q8N6R8 | Gene names | FXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (hFXR1P). | |||||
|
FXR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.078556 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61584, Q8VCU4, Q9R1E2, Q9R1E3, Q9R1E4, Q9R1E5, Q9WUA7, Q9WUA8, Q9WUA9 | Gene names | Fxr1, Fxr1h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (mFxr1p). | |||||
|
C2TA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.014021 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
COFA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.011780 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
FXR2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.074695 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P51116, Q8WUM2 | Gene names | FXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.013435 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
PGCB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.011562 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
PHLB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.008700 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
TDRD3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.046905 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H7E2, Q6P992 | Gene names | TDRD3 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 3. | |||||
|
3BP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.015353 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.019994 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.011753 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
AP4E1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.008421 (rank : 89) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80V94 | Gene names | Ap4e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
CCD71_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.015464 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VEG0, Q3UDX1 | Gene names | Ccdc71 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 71. | |||||
|
CD45_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.003420 (rank : 91) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
DOC10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.007533 (rank : 90) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96BY6, O75178, Q9NW06, Q9NXI8 | Gene names | DOCK10, KIAA0694 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 10 (Protein zizimin 3). | |||||
|
LAP2A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.011391 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61033, Q61028 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms alpha/zeta (Thymopoietin isoforms alpha/zeta) (TP alpha/zeta). | |||||
|
MAVS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.017369 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
NCOA7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.012929 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NI08, Q3LID6, Q4G0V1, Q5TF95, Q6IPQ4, Q6NE83, Q86T89, Q8N1W4 | Gene names | NCOA7, ERAP140, ESNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 7 (140 kDa estrogen receptor-associated protein) (Estrogen nuclear receptor coactivator 1). | |||||
|
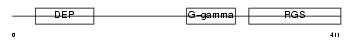
RGS11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.009798 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94810, O75883 | Gene names | RGS11 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 11 (RGS11). | |||||
|
RKHD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.068484 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86XN8 | Gene names | RKHD1, KIAA2031 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger and KH domain-containing protein 1. | |||||
|
VIGLN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.081874 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q00341, Q9UCY3 | Gene names | HDLBP, HBP, VGL | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
VIGLN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.080180 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8VDJ3 | Gene names | Hdlbp | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
AKAP1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
AKAP1_MOUSE
|
||||||
| NC score | 0.938066 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
TDRKH_HUMAN
|
||||||
| NC score | 0.494539 (rank : 3) | θ value | 4.30538e-10 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y2W6, Q8N582, Q9NYV5 | Gene names | TDRKH, TDRD2 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
TDRKH_MOUSE
|
||||||
| NC score | 0.491036 (rank : 4) | θ value | 7.34386e-10 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80VL1 | Gene names | Tdrkh | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
TDRD1_HUMAN
|
||||||
| NC score | 0.425161 (rank : 5) | θ value | 4.0297e-08 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BXT4, Q9H7B3 | Gene names | TDRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
TDRD7_HUMAN
|
||||||
| NC score | 0.423705 (rank : 6) | θ value | 2.79066e-09 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NHU6, Q96JT1, Q9UFF0, Q9Y2M3 | Gene names | TDRD7, PCTAIRE2BP | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 7 (Tudor repeat associator with PCTAIRE 2) (Trap). | |||||
|
TDRD7_MOUSE
|
||||||
| NC score | 0.420218 (rank : 7) | θ value | 8.11959e-09 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K1H1, Q8R181 | Gene names | Tdrd7, Pctaire2bp | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 7 (Tudor repeat associator with PCTAIRE 2) (Trap). | |||||
|
TDRD1_MOUSE
|
||||||
| NC score | 0.405354 (rank : 8) | θ value | 4.45536e-07 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99MV1 | Gene names | Tdrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
TDRD5_HUMAN
|
||||||
| NC score | 0.375762 (rank : 9) | θ value | 0.000121331 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NAT2, Q5VTV0 | Gene names | TDRD5 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 5. | |||||
|
TDRD4_HUMAN
|
||||||
| NC score | 0.338642 (rank : 10) | θ value | 9.29e-05 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NUY9 | Gene names | TDRD4 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 4. | |||||
|
TDRD6_MOUSE
|
||||||
| NC score | 0.274732 (rank : 11) | θ value | 0.0148317 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P61407 | Gene names | Tdrd6 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 6. | |||||
|
NOVA2_HUMAN
|
||||||
| NC score | 0.272156 (rank : 12) | θ value | 0.0563607 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UNW9, O43267, Q9UEA1 | Gene names | NOVA2, ANOVA, NOVA3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-2 (Neuro-oncological ventral antigen 2) (Astrocytic NOVA1-like RNA-binding protein). | |||||
|
NOVA1_HUMAN
|
||||||
| NC score | 0.247379 (rank : 13) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P51513 | Gene names | NOVA1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-1 (Neuro-oncological ventral antigen 1) (Onconeural ventral antigen 1) (Paraneoplastic Ri antigen) (Ventral neuron-specific protein 1). | |||||
|
SND1_HUMAN
|
||||||
| NC score | 0.245638 (rank : 14) | θ value | 0.00390308 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7KZF4, Q13122 | Gene names | SND1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Staphylococcal nuclease domain-containing protein 1 (p100 co- activator) (100 kDa coactivator) (EBNA2 coactivator p100). | |||||
|
FUBP3_HUMAN
|
||||||
| NC score | 0.243925 (rank : 15) | θ value | 0.0113563 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96I24, Q92946, Q9BVB6 | Gene names | FUBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 3 (FUSE-binding protein 3). | |||||
|
HNRPK_HUMAN
|
||||||
| NC score | 0.241076 (rank : 16) | θ value | 0.0193708 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P61978, Q07244, Q15671, Q60577, Q922Y7, Q96J62 | Gene names | HNRPK, HNRNPK | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K (hnRNP K) (Transformation up-regulated nuclear protein) (TUNP). | |||||
|
HNRPK_MOUSE
|
||||||
| NC score | 0.241076 (rank : 17) | θ value | 0.0193708 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P61979, Q07244, Q15671, Q60577, Q8BGQ8, Q922Y7, Q96J62 | Gene names | Hnrpk, Hnrnpk | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K. | |||||
|
SND1_MOUSE
|
||||||
| NC score | 0.231463 (rank : 18) | θ value | 0.0193708 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q78PY7, Q3TT46, Q922L5, Q9R0S1 | Gene names | Snd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Staphylococcal nuclease domain-containing protein 1 (p100 co- activator) (100 kDa coactivator). | |||||
|
PCBP1_HUMAN
|
||||||
| NC score | 0.224697 (rank : 19) | θ value | 0.0330416 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15365, Q13157, Q14975 | Gene names | PCBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) (Nucleic acid- binding protein SUB2.3). | |||||
|
PCBP1_MOUSE
|
||||||
| NC score | 0.224697 (rank : 20) | θ value | 0.0330416 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P60335 | Gene names | Pcbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1). | |||||
|
FUBP2_HUMAN
|
||||||
| NC score | 0.220530 (rank : 21) | θ value | 0.0961366 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92945, O00301, Q9UNT5, Q9UQH5 | Gene names | KHSRP, FUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75). | |||||
|
FUBP1_MOUSE
|
||||||
| NC score | 0.219596 (rank : 22) | θ value | 0.125558 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91WJ8, Q8C0Y8 | Gene names | Fubp1, D3Ertd330e | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP). | |||||
|
FUBP1_HUMAN
|
||||||
| NC score | 0.219184 (rank : 23) | θ value | 0.125558 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96AE4, Q12828 | Gene names | FUBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP) (DNA helicase V) (HDH V). | |||||
|
PCBP3_MOUSE
|
||||||
| NC score | 0.219176 (rank : 24) | θ value | 0.0252991 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P57722, Q8BSB0, Q8C544 | Gene names | Pcbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP3_HUMAN
|
||||||
| NC score | 0.219102 (rank : 25) | θ value | 0.0252991 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P57721, Q8N9K6, Q96EP6 | Gene names | PCBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP2_HUMAN
|
||||||
| NC score | 0.216387 (rank : 26) | θ value | 0.163984 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15366, Q6PKG5 | Gene names | PCBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (hnRNP-E2). | |||||
|
PCBP2_MOUSE
|
||||||
| NC score | 0.215765 (rank : 27) | θ value | 0.163984 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61990, Q61383, Q62042 | Gene names | Pcbp2, Cbp, Hnrnpx, Hnrpx | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP). | |||||
|
PCBP4_MOUSE
|
||||||
| NC score | 0.205586 (rank : 28) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P57724, Q91VR3 | Gene names | Pcbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
PCBP4_HUMAN
|
||||||
| NC score | 0.202396 (rank : 29) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P57723, Q96AH7 | Gene names | PCBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
IF2B2_HUMAN
|
||||||
| NC score | 0.183448 (rank : 30) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y6M1 | Gene names | IGF2BP2, IMP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2) (Hepatocellular carcinoma autoantigen p62). | |||||
|
IF2B2_MOUSE
|
||||||
| NC score | 0.179759 (rank : 31) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5SF07, Q3TCU4, Q7TQF9 | Gene names | Igf2bp2, Imp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2). | |||||
|
PQBP1_MOUSE
|
||||||
| NC score | 0.114033 (rank : 32) | θ value | 0.00869519 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VJ5, Q80WW2, Q9ER43, Q9QYY2 | Gene names | Pqbp1, Npw38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyglutamine-binding protein 1 (Polyglutamine tract-binding protein 1) (PQBP-1) (38 kDa nuclear protein containing a WW domain) (Npw38). | |||||
|
PQBP1_HUMAN
|
||||||
| NC score | 0.090252 (rank : 33) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60828, Q4VY25, Q4VY26, Q4VY27, Q4VY29, Q4VY30, Q4VY34, Q4VY35, Q4VY36, Q4VY37, Q4VY38, Q9GZP2, Q9GZU4, Q9GZZ4 | Gene names | PQBP1, NPW38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyglutamine-binding protein 1 (Polyglutamine tract-binding protein 1) (PQBP-1) (38 kDa nuclear protein containing a WW domain) (Npw38). | |||||
|
VIGLN_HUMAN
|
||||||
| NC score | 0.081874 (rank : 34) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q00341, Q9UCY3 | Gene names | HDLBP, HBP, VGL | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
VIGLN_MOUSE
|
||||||
| NC score | 0.080180 (rank : 35) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8VDJ3 | Gene names | Hdlbp | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
FXR1_HUMAN
|
||||||
| NC score | 0.079409 (rank : 36) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P51114, Q7Z450, Q8N6R8 | Gene names | FXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (hFXR1P). | |||||
|
FXR1_MOUSE
|
||||||
| NC score | 0.078556 (rank : 37) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61584, Q8VCU4, Q9R1E2, Q9R1E3, Q9R1E4, Q9R1E5, Q9WUA7, Q9WUA8, Q9WUA9 | Gene names | Fxr1, Fxr1h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (mFxr1p). | |||||
|
FXR2_HUMAN
|
||||||
| NC score | 0.074695 (rank : 38) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P51116, Q8WUM2 | Gene names | FXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
FMR1_HUMAN
|
||||||
| NC score | 0.069857 (rank : 39) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q06787, Q16578, Q99054 | Gene names | FMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation 1 protein (Protein FMR-1) (FMRP). | |||||
|
FMR1_MOUSE
|
||||||
| NC score | 0.069316 (rank : 40) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35922 | Gene names | Fmr1, Fmr-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation protein 1 homolog (Protein FMR-1) (FMRP) (mFmr1p). | |||||
|
RKHD1_HUMAN
|
||||||
| NC score | 0.068484 (rank : 41) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86XN8 | Gene names | RKHD1, KIAA2031 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger and KH domain-containing protein 1. | |||||
|
CEP41_MOUSE
|
||||||
| NC score | 0.056689 (rank : 42) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99NF3, Q8BT53 | Gene names | Tsga14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 41 kDa (Cep41 protein) (Testis-specific gene A14 protein). | |||||
|
MYOZ3_MOUSE
|
||||||
| NC score | 0.050411 (rank : 43) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R4E4 | Gene names | Myoz3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myozenin-3 (Calsarcin-3) (FATZ-related protein 3). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.049157 (rank : 44) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.048293 (rank : 45) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
TDRD3_HUMAN
|
||||||
| NC score | 0.046905 (rank : 46) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H7E2, Q6P992 | Gene names | TDRD3 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 3. | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.046479 (rank : 47) | θ value | 0.279714 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.045268 (rank : 48) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ANF_MOUSE
|
||||||
| NC score | 0.041389 (rank : 49) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05125 | Gene names | Nppa, Pnd | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic factor precursor (ANF) (Atrial natriuretic peptide) (ANP) (Prepronatriodilatin) [Contains: Auriculin-B; Auriculin-A; Atriopeptin I; Atriopeptin II]. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.040194 (rank : 50) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.039792 (rank : 51) | θ value | 0.0252991 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
PK3CB_HUMAN
|
||||||
| NC score | 0.036526 (rank : 52) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42338 | Gene names | PIK3CB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CB_MOUSE
|
||||||
| NC score | 0.036447 (rank : 53) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BTI9 | Gene names | Pik3cb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.035933 (rank : 54) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
PDLI7_MOUSE
|
||||||
| NC score | 0.035843 (rank : 55) | θ value | 0.163984 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3TJD7, Q80ZY6, Q810S3, Q8C1S4, Q9CRA1 | Gene names | Pdlim7, Enigma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.030833 (rank : 56) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
CN037_HUMAN
|
||||||
| NC score | 0.030582 (rank : 57) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86TY3, Q6P5Q1, Q86TY1 | Gene names | C14orf37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 precursor. | |||||
|
LRRC6_HUMAN
|
||||||
| NC score | 0.028911 (rank : 58) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86X45, Q13648, Q4G183 | Gene names | LRRC6, LRTP, TSLRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 6 (Leucine-rich testis-specific protein) (Testis-specific leucine-rich repeat protein). | |||||
|
AN32B_MOUSE
|
||||||
| NC score | 0.028200 (rank : 59) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
SDPR_MOUSE
|
||||||
| NC score | 0.025831 (rank : 60) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.023209 (rank : 61) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
HSF1_MOUSE
|
||||||
| NC score | 0.022492 (rank : 62) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
K0552_HUMAN
|
||||||
| NC score | 0.021583 (rank : 63) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60299 | Gene names | KIAA0552 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0552. | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.019994 (rank : 64) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
DACH1_MOUSE
|
||||||
| NC score | 0.019351 (rank : 65) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QYB2, O88716, Q8BPQ0, Q8C8D7, Q9QY47, Q9R218, Q9Z0Y5 | Gene names | Dach1, Dach | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.017873 (rank : 66) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
MAVS_HUMAN
|
||||||
| NC score | 0.017369 (rank : 67) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
FRM4A_MOUSE
|
||||||
| NC score | 0.017180 (rank : 68) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BIE6, Q6PDJ4, Q8CHA6 | Gene names | Frmd4a, Frmd4, Kiaa1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FRM4A_HUMAN
|
||||||
| NC score | 0.016730 (rank : 69) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2Q2 | Gene names | FRMD4A, FRMD4, KIAA1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.016337 (rank : 70) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
CCD71_MOUSE
|
||||||
| NC score | 0.015464 (rank : 71) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VEG0, Q3UDX1 | Gene names | Ccdc71 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 71. | |||||
|
HIP1_HUMAN
|
||||||
| NC score | 0.015409 (rank : 72) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
3BP2_HUMAN
|
||||||
| NC score | 0.015353 (rank : 73) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
C2TA_HUMAN
|
||||||
| NC score | 0.014021 (rank : 74) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.014007 (rank : 75) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.013435 (rank : 76) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
NCOA7_HUMAN
|
||||||
| NC score | 0.012929 (rank : 77) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NI08, Q3LID6, Q4G0V1, Q5TF95, Q6IPQ4, Q6NE83, Q86T89, Q8N1W4 | Gene names | NCOA7, ERAP140, ESNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 7 (140 kDa estrogen receptor-associated protein) (Estrogen nuclear receptor coactivator 1). | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.012859 (rank : 78) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
COFA1_HUMAN
|
||||||
| NC score | 0.011780 (rank : 79) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.011753 (rank : 80) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
GOGA5_HUMAN
|
||||||
| NC score | 0.011614 (rank : 81) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
PGCB_MOUSE
|
||||||
| NC score | 0.011562 (rank : 82) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
LAP2A_MOUSE
|
||||||
| NC score | 0.011391 (rank : 83) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61033, Q61028 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms alpha/zeta (Thymopoietin isoforms alpha/zeta) (TP alpha/zeta). | |||||
|
DDX24_HUMAN
|
||||||
| NC score | 0.011229 (rank : 84) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
VWF_MOUSE
|
||||||
| NC score | 0.010615 (rank : 85) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
RGS11_HUMAN
|
||||||
| NC score | 0.009798 (rank : 86) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94810, O75883 | Gene names | RGS11 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 11 (RGS11). | |||||
|
KLK14_HUMAN
|
||||||
| NC score | 0.008842 (rank : 87) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P0G3 | Gene names | KLK14, KLKL6 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-14 precursor (EC 3.4.21.-) (Kallikrein-like protein 6) (KLK-L6). | |||||
|
PHLB2_MOUSE
|
||||||
| NC score | 0.008700 (rank : 88) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
AP4E1_MOUSE
|
||||||
| NC score | 0.008421 (rank : 89) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80V94 | Gene names | Ap4e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
DOC10_HUMAN
|
||||||
| NC score | 0.007533 (rank : 90) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96BY6, O75178, Q9NW06, Q9NXI8 | Gene names | DOCK10, KIAA0694 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 10 (Protein zizimin 3). | |||||
|
CD45_MOUSE
|
||||||
| NC score | 0.003420 (rank : 91) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||