Please be patient as the page loads
|
NCOA7_HUMAN
|
||||||
| SwissProt Accessions | Q8NI08, Q3LID6, Q4G0V1, Q5TF95, Q6IPQ4, Q6NE83, Q86T89, Q8N1W4 | Gene names | NCOA7, ERAP140, ESNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 7 (140 kDa estrogen receptor-associated protein) (Estrogen nuclear receptor coactivator 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NCOA7_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 160 | |
| SwissProt Accessions | Q8NI08, Q3LID6, Q4G0V1, Q5TF95, Q6IPQ4, Q6NE83, Q86T89, Q8N1W4 | Gene names | NCOA7, ERAP140, ESNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 7 (140 kDa estrogen receptor-associated protein) (Estrogen nuclear receptor coactivator 1). | |||||
|
NCOA7_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.952382 (rank : 2) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6DFV7, Q3TAR1, Q3UNB3, Q66K05, Q6ZQM4, Q8BJ15, Q8BJ39 | Gene names | Ncoa7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 7. | |||||
|
OXR1_MOUSE
|
||||||
| θ value | 3.6343e-134 (rank : 3) | NC score | 0.766934 (rank : 4) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
OXR1_HUMAN
|
||||||
| θ value | 2.08775e-89 (rank : 4) | NC score | 0.832806 (rank : 3) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8N573, Q3LIB5, Q6ZVK9, Q8N8V0, Q9H266, Q9NWC7 | Gene names | OXR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1. | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 5) | NC score | 0.122597 (rank : 7) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
SLK_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 6) | NC score | 0.023825 (rank : 135) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 7) | NC score | 0.101196 (rank : 9) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 8) | NC score | 0.067415 (rank : 28) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PEPL_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 9) | NC score | 0.063221 (rank : 37) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 10) | NC score | 0.168190 (rank : 5) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
CHD8_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.025264 (rank : 129) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
MAP2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 12) | NC score | 0.069574 (rank : 23) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
PEPL_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 13) | NC score | 0.051974 (rank : 63) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
AKAP2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 14) | NC score | 0.059425 (rank : 42) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
TRIM6_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 15) | NC score | 0.018986 (rank : 151) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9C030, Q86WZ8, Q9HCR1 | Gene names | TRIM6, RNF89 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 6 (RING finger protein 89). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 0.125558 (rank : 16) | NC score | 0.085726 (rank : 12) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
TRPM6_HUMAN
|
||||||
| θ value | 0.125558 (rank : 17) | NC score | 0.028974 (rank : 117) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BX84, Q6VPR8, Q6VPR9, Q6VPS0, Q6VPS1, Q6VPS2 | Gene names | TRPM6, CHAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
MAP9_HUMAN
|
||||||
| θ value | 0.163984 (rank : 18) | NC score | 0.079981 (rank : 16) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.129346 (rank : 6) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
MACOI_HUMAN
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.053499 (rank : 58) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8N5G2, Q2TLX5, Q2TLX6, Q9NVG6 | Gene names | TMEM57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57). | |||||
|
NOP14_MOUSE
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.074117 (rank : 20) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8R3N1 | Gene names | Nop14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.055900 (rank : 49) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
PCX1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.047568 (rank : 76) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96RV3, O94897, Q96AI7, Q9Y2J9 | Gene names | PCNX, KIAA0805, KIAA0995, PCNXL1 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 1 (Pecanex homolog). | |||||
|
CCAR1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.053482 (rank : 59) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
EFHB_MOUSE
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.039732 (rank : 90) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CDU5 | Gene names | Efhb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.069314 (rank : 25) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.045383 (rank : 82) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
CNGB1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.022042 (rank : 144) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.047014 (rank : 77) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
BMS1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.045586 (rank : 80) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14692, Q86XJ9 | Gene names | BMS1L, KIAA0187 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome biogenesis protein BMS1 homolog. | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.096153 (rank : 10) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
DDX10_MOUSE
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.023217 (rank : 139) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.059611 (rank : 41) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.034408 (rank : 103) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
RPGR_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.051757 (rank : 64) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
DPP4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.028272 (rank : 120) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28843, Q3U514 | Gene names | Dpp4, Cd26 | |||
|
Domain Architecture |

|
|||||
| Description | Dipeptidyl peptidase 4 (EC 3.4.14.5) (Dipeptidyl peptidase IV) (DPP IV) (T-cell activation antigen CD26) (Thymocyte-activating molecule) (THAM) [Contains: Dipeptidyl peptidase 4 membrane form (Dipeptidyl peptidase IV membrane form); Dipeptidyl peptidase 4 soluble form (Dipeptidyl peptidase IV soluble form)]. | |||||
|
GA2L3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.028240 (rank : 121) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86XJ1 | Gene names | GAS2L3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 3 (Growth arrest-specific 2-like 3). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.049017 (rank : 73) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.030392 (rank : 114) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
PML_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.025308 (rank : 128) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60953 | Gene names | Pml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable transcription factor PML. | |||||
|
PP1RA_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.035122 (rank : 102) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80W00, Q811B6, Q8C6T7, Q8K2U8 | Gene names | Ppp1r10, Cat53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (MHC class I region proline-rich protein CAT53). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.048195 (rank : 75) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
SIGL5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.015558 (rank : 156) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15389 | Gene names | SIGLEC5, CD33L2, OBBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid-binding Ig-like lectin 5 precursor (Siglec-5) (Obesity- binding protein 2) (OB-binding protein 2) (OB-BP2) (CD33 antigen-like 2) (CD170 antigen). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.064174 (rank : 36) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
K0146_MOUSE
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.033677 (rank : 104) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BGX7, Q3TCD0, Q5U457, Q6A0B6, Q6IS60, Q811E1, Q8BWD6, Q8R305 | Gene names | Kiaa0146 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0146. | |||||
|
K1210_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.048417 (rank : 74) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
NOP14_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.070625 (rank : 22) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P78316, Q7LGI5, Q7Z6K0, Q8TBR6 | Gene names | NOP14, C4orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
PHLB2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.050115 (rank : 70) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.062097 (rank : 39) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SCN8A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.016694 (rank : 153) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
U2AFM_MOUSE
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.057702 (rank : 45) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q62377 | Gene names | Zrsr2, U2af1-rs2, U2af1l2, U2af1rs2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2). | |||||
|
AHTF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.026775 (rank : 124) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CJF7, Q8BVJ5, Q8VD55 | Gene names | Ahctf1, Elys | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.022534 (rank : 141) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.014135 (rank : 159) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
KI21A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.036910 (rank : 97) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
MERL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.030133 (rank : 115) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P35240, O95683, Q8WUJ2, Q969N0, Q969Q3, Q96T30, Q96T31, Q96T32, Q96T33, Q9BTW3, Q9UNG9, Q9UNH3, Q9UNH4 | Gene names | NF2, SCH | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin) (Schwannomerlin). | |||||
|
MERL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.030006 (rank : 116) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P46662, Q8BR03 | Gene names | Nf2, Nf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin). | |||||
|
PHLB2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.054676 (rank : 53) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
PPR3A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.033656 (rank : 105) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99MR9, Q8BUJ4, Q8BUL0 | Gene names | Ppp1r3a, Pp1g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 3A (Protein phosphatase 1 glycogen-associated regulatory subunit) (Protein phosphatase type-1 glycogen targeting subunit) (RGL). | |||||
|
REV1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.023950 (rank : 133) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBZ9, O95941, Q9C0J4, Q9NUP2 | Gene names | REV1L, REV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein REV1 (EC 2.7.7.-) (Rev1-like terminal deoxycytidyl transferase) (Alpha integrin-binding protein 80) (AIBP80). | |||||
|
RPGR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.054037 (rank : 54) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92834, O00702, O00737, Q8N5T6, Q93039, Q9HD29, Q9UMR1 | Gene names | RPGR, RP3, XLRP3 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator. | |||||
|
SASH1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.022614 (rank : 140) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59808 | Gene names | Sash1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM and SH3 domain-containing protein 1. | |||||
|
U2AFM_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.065851 (rank : 30) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
41_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.030945 (rank : 113) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
EVI1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.002864 (rank : 175) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
LYSM2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.104871 (rank : 8) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IV50, Q5CZ88, Q8WTV3 | Gene names | LYSMD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LysM and putative peptidoglycan-binding domain-containing protein 2. | |||||
|
LYSM2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.091430 (rank : 11) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D7V2, Q80ZR2 | Gene names | Lysmd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LysM and putative peptidoglycan-binding domain-containing protein 2. | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.035754 (rank : 100) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MAP9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.076901 (rank : 18) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.036584 (rank : 98) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.057250 (rank : 46) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
SCN8A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.015955 (rank : 155) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.039973 (rank : 89) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.050806 (rank : 69) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
CIR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.073441 (rank : 21) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
GGA3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.015025 (rank : 158) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
KIF3A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.025389 (rank : 127) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P28741 | Gene names | Kif3a, Kif3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
MACOI_MOUSE
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.046417 (rank : 78) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 620 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7TQE6 | Gene names | Tmem57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57) (Brain-specific adaptor protein C61). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.084199 (rank : 13) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.038663 (rank : 92) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYPT1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.021016 (rank : 148) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
PRS4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.010810 (rank : 167) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62191, P49014, Q03527, Q6IAW0, Q96AZ3 | Gene names | PSMC1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.010810 (rank : 168) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62192, P49014, Q03527, Q96AZ3 | Gene names | Psmc1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
RL1D1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.040137 (rank : 88) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
SNX23_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.027746 (rank : 123) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96L93, Q5HYC0, Q5HYK1, Q5JWW3, Q5TFK5, Q86VL9, Q86YS5, Q8IYU0, Q9BQJ8, Q9BQM0, Q9BQM1, Q9BQM5, Q9H5U0, Q9HCI2, Q9NXN9 | Gene names | C20orf23, KIAA1590, SNX23 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like motor protein C20orf23 (Sorting nexin-23). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.041476 (rank : 86) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
U2AFL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.058204 (rank : 44) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.069424 (rank : 24) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.045419 (rank : 81) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
DPP4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.024454 (rank : 130) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27487 | Gene names | DPP4, ADCP2, CD26 | |||
|
Domain Architecture |

|
|||||
| Description | Dipeptidyl peptidase 4 (EC 3.4.14.5) (Dipeptidyl peptidase IV) (DPP IV) (T-cell activation antigen CD26) (TP103) (Adenosine deaminase complexing protein 2) (ADABP) [Contains: Dipeptidyl peptidase 4 membrane form (Dipeptidyl peptidase IV membrane form); Dipeptidyl peptidase 4 soluble form (Dipeptidyl peptidase IV soluble form)]. | |||||
|
EVI1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.001417 (rank : 178) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 826 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P14404 | Gene names | Evi1, Evi-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
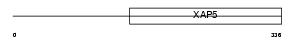
FA50A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.032635 (rank : 109) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9WV03, Q9D866 | Gene names | Fam50a, D0HXS9928E, Xap5 | |||
|
Domain Architecture |
|
|||||
| Description | Protein FAM50A (XAP-5 protein). | |||||
|
FRM4A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.020979 (rank : 149) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BIE6, Q6PDJ4, Q8CHA6 | Gene names | Frmd4a, Frmd4, Kiaa1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.049512 (rank : 71) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.045792 (rank : 79) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
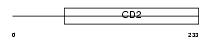
LY9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.015228 (rank : 157) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01965, Q9ES29, Q9ES35, Q9ES36 | Gene names | Ly9, Ly-9 | |||
|
Domain Architecture |
|
|||||
| Description | T-lymphocyte surface antigen Ly-9 precursor (Lymphocyte antigen 9) (Cell-surface molecule Ly-9). | |||||
|
LYSM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.065355 (rank : 33) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D0E3 | Gene names | Lysmd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LysM and putative peptidoglycan-binding domain-containing protein 1. | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.052779 (rank : 60) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.037031 (rank : 96) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
NOP56_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.024205 (rank : 131) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00567, Q9NQ05 | Gene names | NOL5A, NOP56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
STK3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.003572 (rank : 172) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 861 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13188, Q15445, Q15801, Q96FM6 | Gene names | STK3, MST2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 3 (EC 2.7.11.1) (STE20-like kinase MST2) (MST-2) (Mammalian STE20-like protein kinase 2) (Serine/threonine-protein kinase Krs-1). | |||||
|
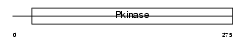
STK3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.003533 (rank : 173) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 869 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JI10, Q60877, Q80UG4, Q8CI58 | Gene names | Stk3, Mess1, Mst2 | |||
|
Domain Architecture |
|
|||||
| Description | Serine/threonine-protein kinase 3 (EC 2.7.11.1) (STE20-like kinase MST2) (MST-2) (Mammalian STE20-like protein kinase 2). | |||||
|
TACC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.037687 (rank : 95) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9JJG0 | Gene names | Tacc2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2. | |||||
|
U2AFL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.053862 (rank : 55) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q64707 | Gene names | Zrsr1, Sp2, Sp2-7, U2af1-rs1, U2af1l1 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1) (SP2). | |||||
|
AKAP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.044643 (rank : 83) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.031710 (rank : 112) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
BCAS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.023843 (rank : 134) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75363, Q68CZ3 | Gene names | BCAS1, AIBC1, NABC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 (Novel amplified in breast cancer 1) (Amplified and overexpressed in breast cancer). | |||||
|
CEP57_MOUSE
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.032748 (rank : 107) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 580 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CEE0, Q6ZQJ3, Q7TN18, Q80X65, Q810F2, Q9D4J4, Q9D5S4, Q9D5W5 | Gene names | Cep57, Kiaa0092, Tsp57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin). | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.016259 (rank : 154) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
FRM4A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.020498 (rank : 150) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P2Q2 | Gene names | FRMD4A, FRMD4, KIAA1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
KMHN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.038368 (rank : 93) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
LMNB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.023251 (rank : 138) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P20700, Q3SYN7, Q96EI6 | Gene names | LMNB1, LMN2, LMNB | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
LMNB1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.023955 (rank : 132) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P14733, Q61791 | Gene names | Lmnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
LYSM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.068458 (rank : 27) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96S90, Q69YX9 | Gene names | LYSMD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LysM and putative peptidoglycan-binding domain-containing protein 1. | |||||
|
MIPO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.035755 (rank : 99) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TD10, Q7Z3J0, Q8IV14 | Gene names | MIPOL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mirror-image polydactyly gene 1 protein. | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.032666 (rank : 108) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.062297 (rank : 38) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NTRK2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.002028 (rank : 176) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15209, Q3TUF9, Q80WU0, Q91XJ9 | Gene names | Ntrk2, Trkb | |||
|
Domain Architecture |

|
|||||
| Description | BDNF/NT-3 growth factors receptor precursor (EC 2.7.10.1) (Neurotrophic tyrosine kinase receptor type 2) (TrkB tyrosine kinase) (GP145-TrkB/GP95-TrkB) (Trk-B). | |||||
|
NUCB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.023611 (rank : 136) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P80303, Q8NFT5 | Gene names | NUCB2, NEFA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-2 precursor (DNA-binding protein NEFA) (Gastric cancer antigen Zg4). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.043069 (rank : 85) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
PP1RA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.028562 (rank : 119) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96QC0, O00405 | Gene names | PPP1R10, CAT53, FB19, PNUTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (Phosphatase 1 nuclear targeting subunit) (MHC class I region proline- rich protein CAT53) (FB19 protein) (PP1-binding protein of 114 kDa) (p99). | |||||
|
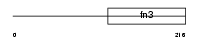
PRLR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.012024 (rank : 164) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08501, P15212, P15213, Q62099 | Gene names | Prlr | |||
|
Domain Architecture |
|
|||||
| Description | Prolactin receptor precursor (PRL-R). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.052425 (rank : 61) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
UBP8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.025510 (rank : 126) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.012732 (rank : 163) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CROP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.049265 (rank : 72) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95232, Q6PHR9, Q9NUY0, Q9P2S7 | Gene names | CROP, CREAP1, O48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein (cAMP regulatory element-associated protein 1) (CRE-associated protein 1) (CREAP-1) (Luc7A) (Okadaic acid-inducible phosphoprotein OA48-18). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.083164 (rank : 14) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
FA50A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.028086 (rank : 122) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14320, Q5HY37 | Gene names | FAM50A, DXS9928E, HXC26, XAP5 | |||
|
Domain Architecture |
|
|||||
| Description | Protein FAM50A (Protein XAP-5) (Protein HXC-26). | |||||
|
IF3A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.038667 (rank : 91) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
IF3A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.038346 (rank : 94) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
LAMC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.013532 (rank : 161) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P11047 | Gene names | LAMC1, LAMB2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
LRC59_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.011583 (rank : 165) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96AG4, Q9P189 | Gene names | LRRC59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 59. | |||||
|
LSP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.023375 (rank : 137) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P33241, Q16096, Q9BUY8 | Gene names | LSP1, WP34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphocyte-specific protein 1 (Protein pp52) (52 kDa phosphoprotein) (Lymphocyte-specific antigen WP34) (47 kDa actin-binding protein). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.011114 (rank : 166) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.033573 (rank : 106) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PININ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.051541 (rank : 66) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.040542 (rank : 87) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
SCN2A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.013925 (rank : 160) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SEC62_MOUSE
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.021716 (rank : 146) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BU14, Q6NX81 | Gene names | Tloc1, Sec62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Translocation protein SEC62 (Translocation protein 1) (TP-1). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.043956 (rank : 84) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
ZN394_HUMAN
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | -0.001218 (rank : 180) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 752 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q53GI3, Q6P5X9, Q8TB27, Q9HA37 | Gene names | ZNF394 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 394. | |||||
|
AKAP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.012929 (rank : 162) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
AT2B1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.003478 (rank : 174) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20020, Q12992, Q12993, Q13819, Q13820, Q13821, Q16504, Q93082 | Gene names | ATP2B1, PMCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 1 (EC 3.6.3.8) (PMCA1) (Plasma membrane calcium pump isoform 1) (Plasma membrane calcium ATPase isoform 1). | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.056800 (rank : 47) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CHD6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.017685 (rank : 152) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
CXA7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.004240 (rank : 171) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P36383 | Gene names | GJA7 | |||
|
Domain Architecture |

|
|||||
| Description | Gap junction alpha-7 protein (Connexin-45) (Cx45). | |||||
|
DNMT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.021720 (rank : 145) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
GATA5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.006685 (rank : 170) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97489 | Gene names | Gata5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
HMCN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | -0.000095 (rank : 179) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 983 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96RW7, Q5TYR7, Q96DN3, Q96DN8, Q96SC3 | Gene names | HMCN1, FIBL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemicentin-1 precursor (Fibulin-6) (FIBL-6). | |||||
|
MAP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.064703 (rank : 35) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.028779 (rank : 118) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.031862 (rank : 110) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
NTBP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.021457 (rank : 147) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5T7V8, Q49A22, Q6P1P9, Q9HAE6, Q9Y350 | Gene names | SCYL1BP1, NTKLBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like-binding protein 1 (NTKL-binding protein 1) (NTKL-BP1) (hNTKL-BP1) (SCY1-like 1-binding protein 1) (SCYL1-binding protein 1) (SCYL1-BP1). | |||||
|
NTRK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.001577 (rank : 177) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16620, Q16675, Q8WXJ6 | Gene names | NTRK2, TRKB | |||
|
Domain Architecture |

|
|||||
| Description | BDNF/NT-3 growth factors receptor precursor (EC 2.7.10.1) (Neurotrophic tyrosine kinase receptor type 2) (TrkB tyrosine kinase) (GP145-TrkB) (Trk-B). | |||||
|
NUCB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.025527 (rank : 125) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P81117 | Gene names | Nucb2, Nefa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-2 precursor (DNA-binding protein NEFA). | |||||
|
SLK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.022291 (rank : 143) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.035387 (rank : 101) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TRI66_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.007643 (rank : 169) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
ZCH10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.031751 (rank : 111) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
ZN638_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.022320 (rank : 142) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.077875 (rank : 17) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.053505 (rank : 57) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
CI039_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.055451 (rank : 51) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.082032 (rank : 15) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
FILA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.055974 (rank : 48) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
GON4L_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.065402 (rank : 32) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.066244 (rank : 29) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.051336 (rank : 67) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.061759 (rank : 40) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MATR3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.052198 (rank : 62) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
NASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.051231 (rank : 68) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.074442 (rank : 19) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.054725 (rank : 52) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.055642 (rank : 50) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
RSRC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.065520 (rank : 31) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96IZ7, Q96QK2, Q9NZE5 | Gene names | RSRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich coiled coil protein 1. | |||||
|
RSRC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.068729 (rank : 26) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DBU6, Q3TF06 | Gene names | Rsrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich coiled coil protein 1. | |||||
|
SIAL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.059282 (rank : 43) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P21815 | Gene names | IBSP, BNSP | |||
|
Domain Architecture |
|
|||||
| Description | Bone sialoprotein-2 precursor (Bone sialoprotein II) (BSP II) (Cell- binding sialoprotein) (Integrin-binding sialoprotein). | |||||
|
TTC16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.053748 (rank : 56) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.051568 (rank : 65) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.065060 (rank : 34) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
NCOA7_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 160 | |
| SwissProt Accessions | Q8NI08, Q3LID6, Q4G0V1, Q5TF95, Q6IPQ4, Q6NE83, Q86T89, Q8N1W4 | Gene names | NCOA7, ERAP140, ESNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 7 (140 kDa estrogen receptor-associated protein) (Estrogen nuclear receptor coactivator 1). | |||||
|
NCOA7_MOUSE
|
||||||
| NC score | 0.952382 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6DFV7, Q3TAR1, Q3UNB3, Q66K05, Q6ZQM4, Q8BJ15, Q8BJ39 | Gene names | Ncoa7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 7. | |||||
|
OXR1_HUMAN
|
||||||
| NC score | 0.832806 (rank : 3) | θ value | 2.08775e-89 (rank : 4) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8N573, Q3LIB5, Q6ZVK9, Q8N8V0, Q9H266, Q9NWC7 | Gene names | OXR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1. | |||||
|
OXR1_MOUSE
|
||||||
| NC score | 0.766934 (rank : 4) | θ value | 3.6343e-134 (rank : 3) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.168190 (rank : 5) | θ value | 0.0431538 (rank : 10) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.129346 (rank : 6) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.122597 (rank : 7) | θ value | 0.00390308 (rank : 5) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
LYSM2_HUMAN
|
||||||
| NC score | 0.104871 (rank : 8) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IV50, Q5CZ88, Q8WTV3 | Gene names | LYSMD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LysM and putative peptidoglycan-binding domain-containing protein 2. | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.101196 (rank : 9) | θ value | 0.00665767 (rank : 7) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.096153 (rank : 10) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
LYSM2_MOUSE
|
||||||
| NC score | 0.091430 (rank : 11) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D7V2, Q80ZR2 | Gene names | Lysmd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LysM and putative peptidoglycan-binding domain-containing protein 2. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.085726 (rank : 12) | θ value | 0.125558 (rank : 16) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.084199 (rank : 13) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.083164 (rank : 14) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.082032 (rank : 15) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MAP9_HUMAN
|
||||||
| NC score | 0.079981 (rank : 16) | θ value | 0.163984 (rank : 18) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.077875 (rank : 17) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
MAP9_MOUSE
|
||||||
| NC score | 0.076901 (rank : 18) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.074442 (rank : 19) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
NOP14_MOUSE
|
||||||
| NC score | 0.074117 (rank : 20) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8R3N1 | Gene names | Nop14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
CIR_HUMAN
|
||||||
| NC score | 0.073441 (rank : 21) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
NOP14_HUMAN
|
||||||
| NC score | 0.070625 (rank : 22) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P78316, Q7LGI5, Q7Z6K0, Q8TBR6 | Gene names | NOP14, C4orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
MAP2_HUMAN
|
||||||
| NC score | 0.069574 (rank : 23) | θ value | 0.0563607 (rank : 12) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.069424 (rank : 24) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.069314 (rank : 25) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
RSRC1_MOUSE
|
||||||
| NC score | 0.068729 (rank : 26) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DBU6, Q3TF06 | Gene names | Rsrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich coiled coil protein 1. | |||||
|
LYSM1_HUMAN
|
||||||
| NC score | 0.068458 (rank : 27) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96S90, Q69YX9 | Gene names | LYSMD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LysM and putative peptidoglycan-binding domain-containing protein 1. | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.067415 (rank : 28) | θ value | 0.0148317 (rank : 8) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.066244 (rank : 29) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
U2AFM_HUMAN
|
||||||
| NC score | 0.065851 (rank : 30) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
RSRC1_HUMAN
|
||||||
| NC score | 0.065520 (rank : 31) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96IZ7, Q96QK2, Q9NZE5 | Gene names | RSRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich coiled coil protein 1. | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.065402 (rank : 32) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
LYSM1_MOUSE
|
||||||
| NC score | 0.065355 (rank : 33) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D0E3 | Gene names | Lysmd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LysM and putative peptidoglycan-binding domain-containing protein 1. | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.065060 (rank : 34) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
MAP2_MOUSE
|
||||||
| NC score | 0.064703 (rank : 35) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.064174 (rank : 36) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
PEPL_MOUSE
|
||||||
| NC score | 0.063221 (rank : 37) | θ value | 0.0193708 (rank : 9) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.062297 (rank : 38) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.062097 (rank : 39) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.061759 (rank : 40) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.059611 (rank : 41) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
AKAP2_HUMAN
|
||||||
| NC score | 0.059425 (rank : 42) | θ value | 0.0961366 (rank : 14) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
SIAL_HUMAN
|
||||||
| NC score | 0.059282 (rank : 43) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P21815 | Gene names | IBSP, BNSP | |||
|
Domain Architecture |

|
|||||
| Description | Bone sialoprotein-2 precursor (Bone sialoprotein II) (BSP II) (Cell- binding sialoprotein) (Integrin-binding sialoprotein). | |||||
|
U2AFL_HUMAN
|
||||||
| NC score | 0.058204 (rank : 44) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
U2AFM_MOUSE
|
||||||
| NC score | 0.057702 (rank : 45) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q62377 | Gene names | Zrsr2, U2af1-rs2, U2af1l2, U2af1rs2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.057250 (rank : 46) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.056800 (rank : 47) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.055974 (rank : 48) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.055900 (rank : 49) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.055642 (rank : 50) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.055451 (rank : 51) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.054725 (rank : 52) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
PHLB2_MOUSE
|
||||||
| NC score | 0.054676 (rank : 53) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
RPGR_HUMAN
|
||||||
| NC score | 0.054037 (rank : 54) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92834, O00702, O00737, Q8N5T6, Q93039, Q9HD29, Q9UMR1 | Gene names | RPGR, RP3, XLRP3 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator. | |||||
|
U2AFL_MOUSE
|
||||||
| NC score | 0.053862 (rank : 55) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q64707 | Gene names | Zrsr1, Sp2, Sp2-7, U2af1-rs1, U2af1l1 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1) (SP2). | |||||
|
TTC16_HUMAN
|
||||||
| NC score | 0.053748 (rank : 56) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.053505 (rank : 57) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
MACOI_HUMAN
|
||||||
| NC score | 0.053499 (rank : 58) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8N5G2, Q2TLX5, Q2TLX6, Q9NVG6 | Gene names | TMEM57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57). | |||||
|
CCAR1_HUMAN
|
||||||
| NC score | 0.053482 (rank : 59) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.052779 (rank : 60) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.052425 (rank : 61) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.052198 (rank : 62) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.051974 (rank : 63) | θ value | 0.0736092 (rank : 13) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
RPGR_MOUSE
|
||||||
| NC score | 0.051757 (rank : 64) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.051568 (rank : 65) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.051541 (rank : 66) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.051336 (rank : 67) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.051231 (rank : 68) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.050806 (rank : 69) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
PHLB2_HUMAN
|
||||||
| NC score | 0.050115 (rank : 70) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.049512 (rank : 71) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
CROP_HUMAN
|
||||||
| NC score | 0.049265 (rank : 72) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95232, Q6PHR9, Q9NUY0, Q9P2S7 | Gene names | CROP, CREAP1, O48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein (cAMP regulatory element-associated protein 1) (CRE-associated protein 1) (CREAP-1) (Luc7A) (Okadaic acid-inducible phosphoprotein OA48-18). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.049017 (rank : 73) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
K1210_HUMAN
|
||||||
| NC score | 0.048417 (rank : 74) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.048195 (rank : 75) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
PCX1_HUMAN
|
||||||
| NC score | 0.047568 (rank : 76) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96RV3, O94897, Q96AI7, Q9Y2J9 | Gene names | PCNX, KIAA0805, KIAA0995, PCNXL1 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 1 (Pecanex homolog). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.047014 (rank : 77) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
MACOI_MOUSE
|
||||||
| NC score | 0.046417 (rank : 78) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 620 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7TQE6 | Gene names | Tmem57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57) (Brain-specific adaptor protein C61). | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.045792 (rank : 79) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
BMS1_HUMAN
|
||||||
| NC score | 0.045586 (rank : 80) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14692, Q86XJ9 | Gene names | BMS1L, KIAA0187 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome biogenesis protein BMS1 homolog. | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.045419 (rank : 81) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.045383 (rank : 82) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
AKAP2_MOUSE
|
||||||
| NC score | 0.044643 (rank : 83) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.043956 (rank : 84) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.043069 (rank : 85) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.041476 (rank : 86) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.040542 (rank : 87) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
RL1D1_HUMAN
|
||||||
| NC score | 0.040137 (rank : 88) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.039973 (rank : 89) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
EFHB_MOUSE
|
||||||
| NC score | 0.039732 (rank : 90) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CDU5 | Gene names | Efhb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
IF3A_HUMAN
|
||||||
| NC score | 0.038667 (rank : 91) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.038663 (rank : 92) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
KMHN1_MOUSE
|
||||||
| NC score | 0.038368 (rank : 93) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
IF3A_MOUSE
|
||||||
| NC score | 0.038346 (rank : 94) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
TACC2_MOUSE
|
||||||
| NC score | 0.037687 (rank : 95) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9JJG0 | Gene names | Tacc2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2. | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.037031 (rank : 96) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
KI21A_HUMAN
|
||||||
| NC score | 0.036910 (rank : 97) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.036584 (rank : 98) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MIPO1_HUMAN
|
||||||
| NC score | 0.035755 (rank : 99) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TD10, Q7Z3J0, Q8IV14 | Gene names | MIPOL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mirror-image polydactyly gene 1 protein. | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.035754 (rank : 100) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.035387 (rank : 101) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
PP1RA_MOUSE
|
||||||
| NC score | 0.035122 (rank : 102) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80W00, Q811B6, Q8C6T7, Q8K2U8 | Gene names | Ppp1r10, Cat53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (MHC class I region proline-rich protein CAT53). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.034408 (rank : 103) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
K0146_MOUSE
|
||||||
| NC score | 0.033677 (rank : 104) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BGX7, Q3TCD0, Q5U457, Q6A0B6, Q6IS60, Q811E1, Q8BWD6, Q8R305 | Gene names | Kiaa0146 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0146. | |||||
|
PPR3A_MOUSE
|
||||||
| NC score | 0.033656 (rank : 105) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99MR9, Q8BUJ4, Q8BUL0 | Gene names | Ppp1r3a, Pp1g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 3A (Protein phosphatase 1 glycogen-associated regulatory subunit) (Protein phosphatase type-1 glycogen targeting subunit) (RGL). | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.033573 (rank : 106) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
CEP57_MOUSE
|
||||||
| NC score | 0.032748 (rank : 107) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 580 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CEE0, Q6ZQJ3, Q7TN18, Q80X65, Q810F2, Q9D4J4, Q9D5S4, Q9D5W5 | Gene names | Cep57, Kiaa0092, Tsp57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.032666 (rank : 108) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
FA50A_MOUSE
|
||||||
| NC score | 0.032635 (rank : 109) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9WV03, Q9D866 | Gene names | Fam50a, D0HXS9928E, Xap5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein FAM50A (XAP-5 protein). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.031862 (rank : 110) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
ZCH10_MOUSE
|
||||||
| NC score | 0.031751 (rank : 111) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.031710 (rank : 112) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
41_HUMAN
|
||||||
| NC score | 0.030945 (rank : 113) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
KI21A_MOUSE
|
||||||
| NC score | 0.030392 (rank : 114) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
MERL_HUMAN
|
||||||
| NC score | 0.030133 (rank : 115) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P35240, O95683, Q8WUJ2, Q969N0, Q969Q3, Q96T30, Q96T31, Q96T32, Q96T33, Q9BTW3, Q9UNG9, Q9UNH3, Q9UNH4 | Gene names | NF2, SCH | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin) (Schwannomerlin). | |||||
|
MERL_MOUSE
|
||||||
| NC score | 0.030006 (rank : 116) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P46662, Q8BR03 | Gene names | Nf2, Nf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin). | |||||
|
TRPM6_HUMAN
|
||||||
| NC score | 0.028974 (rank : 117) | θ value | 0.125558 (rank : 17) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BX84, Q6VPR8, Q6VPR9, Q6VPS0, Q6VPS1, Q6VPS2 | Gene names | TRPM6, CHAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.028779 (rank : 118) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
PP1RA_HUMAN
|
||||||
| NC score | 0.028562 (rank : 119) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96QC0, O00405 | Gene names | PPP1R10, CAT53, FB19, PNUTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (Phosphatase 1 nuclear targeting subunit) (MHC class I region proline- rich protein CAT53) (FB19 protein) (PP1-binding protein of 114 kDa) (p99). | |||||
|
DPP4_MOUSE
|
||||||
| NC score | 0.028272 (rank : 120) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28843, Q3U514 | Gene names | Dpp4, Cd26 | |||
|
Domain Architecture |

|
|||||
| Description | Dipeptidyl peptidase 4 (EC 3.4.14.5) (Dipeptidyl peptidase IV) (DPP IV) (T-cell activation antigen CD26) (Thymocyte-activating molecule) (THAM) [Contains: Dipeptidyl peptidase 4 membrane form (Dipeptidyl peptidase IV membrane form); Dipeptidyl peptidase 4 soluble form (Dipeptidyl peptidase IV soluble form)]. | |||||
|
GA2L3_HUMAN
|
||||||
| NC score | 0.028240 (rank : 121) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86XJ1 | Gene names | GAS2L3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 3 (Growth arrest-specific 2-like 3). | |||||
|
FA50A_HUMAN
|
||||||
| NC score | 0.028086 (rank : 122) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14320, Q5HY37 | Gene names | FAM50A, DXS9928E, HXC26, XAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein FAM50A (Protein XAP-5) (Protein HXC-26). | |||||
|
SNX23_HUMAN
|
||||||
| NC score | 0.027746 (rank : 123) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96L93, Q5HYC0, Q5HYK1, Q5JWW3, Q5TFK5, Q86VL9, Q86YS5, Q8IYU0, Q9BQJ8, Q9BQM0, Q9BQM1, Q9BQM5, Q9H5U0, Q9HCI2, Q9NXN9 | Gene names | C20orf23, KIAA1590, SNX23 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like motor protein C20orf23 (Sorting nexin-23). | |||||
|
AHTF1_MOUSE
|
||||||
| NC score | 0.026775 (rank : 124) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CJF7, Q8BVJ5, Q8VD55 | Gene names | Ahctf1, Elys | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
NUCB2_MOUSE
|
||||||
| NC score | 0.025527 (rank : 125) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P81117 | Gene names | Nucb2, Nefa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-2 precursor (DNA-binding protein NEFA). | |||||
|
UBP8_HUMAN
|
||||||
| NC score | 0.025510 (rank : 126) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
KIF3A_MOUSE
|
||||||
| NC score | 0.025389 (rank : 127) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P28741 | Gene names | Kif3a, Kif3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
PML_MOUSE
|
||||||
| NC score | 0.025308 (rank : 128) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60953 | Gene names | Pml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable transcription factor PML. | |||||
|
CHD8_HUMAN
|
||||||
| NC score | 0.025264 (rank : 129) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
DPP4_HUMAN
|
||||||
| NC score | 0.024454 (rank : 130) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27487 | Gene names | DPP4, ADCP2, CD26 | |||
|
Domain Architecture |

|
|||||
| Description | Dipeptidyl peptidase 4 (EC 3.4.14.5) (Dipeptidyl peptidase IV) (DPP IV) (T-cell activation antigen CD26) (TP103) (Adenosine deaminase complexing protein 2) (ADABP) [Contains: Dipeptidyl peptidase 4 membrane form (Dipeptidyl peptidase IV membrane form); Dipeptidyl peptidase 4 soluble form (Dipeptidyl peptidase IV soluble form)]. | |||||
|
NOP56_HUMAN
|
||||||
| NC score | 0.024205 (rank : 131) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00567, Q9NQ05 | Gene names | NOL5A, NOP56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
LMNB1_MOUSE
|
||||||
| NC score | 0.023955 (rank : 132) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P14733, Q61791 | Gene names | Lmnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
REV1_HUMAN
|
||||||
| NC score | 0.023950 (rank : 133) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBZ9, O95941, Q9C0J4, Q9NUP2 | Gene names | REV1L, REV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein REV1 (EC 2.7.7.-) (Rev1-like terminal deoxycytidyl transferase) (Alpha integrin-binding protein 80) (AIBP80). | |||||
|
BCAS1_HUMAN
|
||||||
| NC score | 0.023843 (rank : 134) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75363, Q68CZ3 | Gene names | BCAS1, AIBC1, NABC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 (Novel amplified in breast cancer 1) (Amplified and overexpressed in breast cancer). | |||||
|
SLK_HUMAN
|
||||||
| NC score | 0.023825 (rank : 135) | θ value | 0.00390308 (rank : 6) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
NUCB2_HUMAN
|
||||||
| NC score | 0.023611 (rank : 136) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P80303, Q8NFT5 | Gene names | NUCB2, NEFA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-2 precursor (DNA-binding protein NEFA) (Gastric cancer antigen Zg4). | |||||
|
LSP1_HUMAN
|
||||||
| NC score | 0.023375 (rank : 137) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P33241, Q16096, Q9BUY8 | Gene names | LSP1, WP34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphocyte-specific protein 1 (Protein pp52) (52 kDa phosphoprotein) (Lymphocyte-specific antigen WP34) (47 kDa actin-binding protein). | |||||
|
LMNB1_HUMAN
|
||||||
| NC score | 0.023251 (rank : 138) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P20700, Q3SYN7, Q96EI6 | Gene names | LMNB1, LMN2, LMNB | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
DDX10_MOUSE
|
||||||
| NC score | 0.023217 (rank : 139) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
SASH1_MOUSE
|
||||||
| NC score | 0.022614 (rank : 140) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59808 | Gene names | Sash1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM and SH3 domain-containing protein 1. | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.022534 (rank : 141) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ZN638_MOUSE
|
||||||
| NC score | 0.022320 (rank : 142) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
SLK_MOUSE
|
||||||
| NC score | 0.022291 (rank : 143) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
CNGB1_HUMAN
|
||||||
| NC score | 0.022042 (rank : 144) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
DNMT1_HUMAN
|
||||||
| NC score | 0.021720 (rank : 145) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
SEC62_MOUSE
|
||||||
| NC score | 0.021716 (rank : 146) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BU14, Q6NX81 | Gene names | Tloc1, Sec62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Translocation protein SEC62 (Translocation protein 1) (TP-1). | |||||
|
NTBP1_HUMAN
|
||||||
| NC score | 0.021457 (rank : 147) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5T7V8, Q49A22, Q6P1P9, Q9HAE6, Q9Y350 | Gene names | SCYL1BP1, NTKLBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like-binding protein 1 (NTKL-binding protein 1) (NTKL-BP1) (hNTKL-BP1) (SCY1-like 1-binding protein 1) (SCYL1-binding protein 1) (SCYL1-BP1). | |||||
|
MYPT1_MOUSE
|
||||||
| NC score | 0.021016 (rank : 148) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
FRM4A_MOUSE
|
||||||
| NC score | 0.020979 (rank : 149) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BIE6, Q6PDJ4, Q8CHA6 | Gene names | Frmd4a, Frmd4, Kiaa1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FRM4A_HUMAN
|
||||||
| NC score | 0.020498 (rank : 150) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P2Q2 | Gene names | FRMD4A, FRMD4, KIAA1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
TRIM6_HUMAN
|
||||||
| NC score | 0.018986 (rank : 151) | θ value | 0.0961366 (rank : 15) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9C030, Q86WZ8, Q9HCR1 | Gene names | TRIM6, RNF89 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 6 (RING finger protein 89). | |||||
|
CHD6_HUMAN
|
||||||
| NC score | 0.017685 (rank : 152) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
SCN8A_MOUSE
|
||||||
| NC score | 0.016694 (rank : 153) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.016259 (rank : 154) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
SCN8A_HUMAN
|
||||||
| NC score | 0.015955 (rank : 155) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SIGL5_HUMAN
|
||||||
| NC score | 0.015558 (rank : 156) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15389 | Gene names | SIGLEC5, CD33L2, OBBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid-binding Ig-like lectin 5 precursor (Siglec-5) (Obesity- binding protein 2) (OB-binding protein 2) (OB-BP2) (CD33 antigen-like 2) (CD170 antigen). | |||||
|
LY9_MOUSE
|
||||||
| NC score | 0.015228 (rank : 157) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01965, Q9ES29, Q9ES35, Q9ES36 | Gene names | Ly9, Ly-9 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphocyte surface antigen Ly-9 precursor (Lymphocyte antigen 9) (Cell-surface molecule Ly-9). | |||||
|
GGA3_MOUSE
|
||||||
| NC score | 0.015025 (rank : 158) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.014135 (rank : 159) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
SCN2A_HUMAN
|
||||||
| NC score | 0.013925 (rank : 160) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
LAMC1_HUMAN
|
||||||
| NC score | 0.013532 (rank : 161) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P11047 | Gene names | LAMC1, LAMB2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
AKAP1_HUMAN
|
||||||
| NC score | 0.012929 (rank : 162) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.012732 (rank : 163) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
PRLR_MOUSE
|
||||||
| NC score | 0.012024 (rank : 164) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08501, P15212, P15213, Q62099 | Gene names | Prlr | |||
|
Domain Architecture |

|
|||||
| Description | Prolactin receptor precursor (PRL-R). | |||||
|
LRC59_HUMAN
|
||||||
| NC score | 0.011583 (rank : 165) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96AG4, Q9P189 | Gene names | LRRC59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 59. | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.011114 (rank : 166) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
PRS4_HUMAN
|
||||||
| NC score | 0.010810 (rank : 167) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62191, P49014, Q03527, Q6IAW0, Q96AZ3 | Gene names | PSMC1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
PRS4_MOUSE
|
||||||
| NC score | 0.010810 (rank : 168) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62192, P49014, Q03527, Q96AZ3 | Gene names | Psmc1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit 4 (P26s4) (Proteasome 26S subunit ATPase 1). | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.007643 (rank : 169) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
GATA5_MOUSE
|
||||||
| NC score | 0.006685 (rank : 170) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97489 | Gene names | Gata5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
CXA7_HUMAN
|
||||||
| NC score | 0.004240 (rank : 171) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P36383 | Gene names | GJA7 | |||
|
Domain Architecture |

|
|||||
| Description | Gap junction alpha-7 protein (Connexin-45) (Cx45). | |||||
|
STK3_HUMAN
|
||||||
| NC score | 0.003572 (rank : 172) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 861 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13188, Q15445, Q15801, Q96FM6 | Gene names | STK3, MST2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 3 (EC 2.7.11.1) (STE20-like kinase MST2) (MST-2) (Mammalian STE20-like protein kinase 2) (Serine/threonine-protein kinase Krs-1). | |||||
|
STK3_MOUSE
|
||||||
| NC score | 0.003533 (rank : 173) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 869 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JI10, Q60877, Q80UG4, Q8CI58 | Gene names | Stk3, Mess1, Mst2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 3 (EC 2.7.11.1) (STE20-like kinase MST2) (MST-2) (Mammalian STE20-like protein kinase 2). | |||||
|
AT2B1_HUMAN
|
||||||
| NC score | 0.003478 (rank : 174) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20020, Q12992, Q12993, Q13819, Q13820, Q13821, Q16504, Q93082 | Gene names | ATP2B1, PMCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 1 (EC 3.6.3.8) (PMCA1) (Plasma membrane calcium pump isoform 1) (Plasma membrane calcium ATPase isoform 1). | |||||
|
EVI1_HUMAN
|
||||||
| NC score | 0.002864 (rank : 175) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
NTRK2_MOUSE
|
||||||
| NC score | 0.002028 (rank : 176) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15209, Q3TUF9, Q80WU0, Q91XJ9 | Gene names | Ntrk2, Trkb | |||
|
Domain Architecture |

|
|||||
| Description | BDNF/NT-3 growth factors receptor precursor (EC 2.7.10.1) (Neurotrophic tyrosine kinase receptor type 2) (TrkB tyrosine kinase) (GP145-TrkB/GP95-TrkB) (Trk-B). | |||||
|
NTRK2_HUMAN
|
||||||
| NC score | 0.001577 (rank : 177) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16620, Q16675, Q8WXJ6 | Gene names | NTRK2, TRKB | |||
|
Domain Architecture |

|
|||||
| Description | BDNF/NT-3 growth factors receptor precursor (EC 2.7.10.1) (Neurotrophic tyrosine kinase receptor type 2) (TrkB tyrosine kinase) (GP145-TrkB) (Trk-B). | |||||
|
EVI1_MOUSE
|
||||||
| NC score | 0.001417 (rank : 178) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 826 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P14404 | Gene names | Evi1, Evi-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
HMCN1_HUMAN
|
||||||
| NC score | -0.000095 (rank : 179) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 983 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96RW7, Q5TYR7, Q96DN3, Q96DN8, Q96SC3 | Gene names | HMCN1, FIBL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemicentin-1 precursor (Fibulin-6) (FIBL-6). | |||||
|
ZN394_HUMAN
|
||||||
| NC score | -0.001218 (rank : 180) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 752 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q53GI3, Q6P5X9, Q8TB27, Q9HA37 | Gene names | ZNF394 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 394. | |||||