Please be patient as the page loads
|
DOC10_HUMAN
|
||||||
| SwissProt Accessions | Q96BY6, O75178, Q9NW06, Q9NXI8 | Gene names | DOCK10, KIAA0694 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 10 (Protein zizimin 3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DOC10_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q96BY6, O75178, Q9NW06, Q9NXI8 | Gene names | DOCK10, KIAA0694 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 10 (Protein zizimin 3). | |||||
|
DOCK9_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.977872 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BZ29, Q5JUD4, Q5JUD6, Q5T2Q1, Q5TAN8, Q9BZ25, Q9BZ26, Q9BZ27, Q9BZ28, Q9UPU4 | Gene names | DOCK9, KIAA1058 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 9 (Cdc42 guanine nucleotide exchange factor zizimin 1). | |||||
|
DOCK9_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.978787 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BIK4, Q921Y6 | Gene names | Dock9, D14Wsu89e, Kiaa1058 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 9 (Cdc42 guanine nucleotide exchange factor zizimin 1). | |||||
|
DOCK7_HUMAN
|
||||||
| θ value | 4.72459e-150 (rank : 4) | NC score | 0.913205 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96N67, Q6ZV32, Q8TB82, Q96NG6, Q9C092 | Gene names | DOCK7, KIAA1771 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dedicator of cytokinesis protein 7. | |||||
|
DOCK6_HUMAN
|
||||||
| θ value | 1.28961e-139 (rank : 5) | NC score | 0.912800 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96HP0, Q9P2F2 | Gene names | DOCK6, KIAA1395 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 6. | |||||
|
DOCK8_HUMAN
|
||||||
| θ value | 3.51195e-137 (rank : 6) | NC score | 0.901250 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NF50, Q3MV16, Q5JPJ1, Q8TEP1, Q8WUY2, Q9BYJ5, Q9H1Q2, Q9H1Q3, Q9H308, Q9H7P2 | Gene names | DOCK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dedicator of cytokinesis protein 8. | |||||
|
DOCK8_MOUSE
|
||||||
| θ value | 4.58675e-137 (rank : 7) | NC score | 0.912780 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C147, Q6PIS0, Q7TMQ5, Q8K105, Q9DBQ2 | Gene names | Dock8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dedicator of cytokinesis protein 8. | |||||
|
DOCK3_HUMAN
|
||||||
| θ value | 1.13037e-18 (rank : 8) | NC score | 0.492703 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IZD9, O15017 | Gene names | DOCK3, KIAA0299, MOCA | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 3 (Modifier of cell adhesion) (Presenilin-binding protein) (PBP protein). | |||||
|
DOCK3_MOUSE
|
||||||
| θ value | 3.28887e-18 (rank : 9) | NC score | 0.492096 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CIQ7 | Gene names | Dock3, Moca | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 3 (Modifier of cell adhesion) (Presenilin-binding protein) (PBP protein). | |||||
|
DOCK2_MOUSE
|
||||||
| θ value | 6.85773e-16 (rank : 10) | NC score | 0.448597 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C3J5, Q99M79 | Gene names | Dock2 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 2 (Hch protein). | |||||
|
DOCK2_HUMAN
|
||||||
| θ value | 1.52774e-15 (rank : 11) | NC score | 0.449803 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92608, Q96AK7 | Gene names | DOCK2, KIAA0209 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 2. | |||||
|
DOCK4_HUMAN
|
||||||
| θ value | 5.8054e-15 (rank : 12) | NC score | 0.457602 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK4_MOUSE
|
||||||
| θ value | 9.90251e-15 (rank : 13) | NC score | 0.456588 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK1_HUMAN
|
||||||
| θ value | 8.38298e-14 (rank : 14) | NC score | 0.435049 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14185 | Gene names | DOCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 1 (180 kDa protein downstream of CRK) (DOCK180). | |||||
|
GAB2_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 15) | NC score | 0.141770 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
GAB2_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 16) | NC score | 0.153334 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UQC2, O60317 | Gene names | GAB2, KIAA0571 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (pp100). | |||||
|
RHG25_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 17) | NC score | 0.115310 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG25_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 18) | NC score | 0.103007 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 19) | NC score | 0.031343 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 20) | NC score | 0.050195 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 21) | NC score | 0.063153 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
GAB1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 22) | NC score | 0.121136 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
PKHA5_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 23) | NC score | 0.105218 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 24) | NC score | 0.025478 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 25) | NC score | 0.111351 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 0.163984 (rank : 26) | NC score | 0.022247 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 0.163984 (rank : 27) | NC score | 0.069328 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.021429 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
IFT81_MOUSE
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.024687 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
NU107_HUMAN
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.044035 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57740 | Gene names | NUP107 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup107 (Nucleoporin Nup107) (107 kDa nucleoporin). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.019580 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
RPC1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.023881 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14802, Q8TCW5 | Gene names | POLR3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase III largest subunit (EC 2.7.7.6) (RPC155) (RPC1). | |||||
|
PKHA1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.060557 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PLEK2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.075688 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NYT0, Q96JT0 | Gene names | PLEK2 | |||
|
Domain Architecture |
|
|||||
| Description | Pleckstrin-2. | |||||
|
PLEK2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.069338 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WV52 | Gene names | Plek2 | |||
|
Domain Architecture |
|
|||||
| Description | Pleckstrin-2. | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.019250 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
LAMB1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.012900 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
IRS2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.030170 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
MPP2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.009698 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WV34 | Gene names | Mpp2, Dlgh2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 2 (Protein MPP2) (Dlgh2 protein). | |||||
|
NU107_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.038416 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BH74, Q99KH5 | Gene names | Nup107 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup107 (Nucleoporin Nup107) (107 kDa nucleoporin). | |||||
|
PLEK_HUMAN
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.090924 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.026853 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
TRI14_HUMAN
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.014299 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14142, Q9BRD8, Q9C020 | Gene names | TRIM14, KIAA0129 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 14. | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.014002 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
DP13A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.023818 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UKG1, Q9P2B9 | Gene names | APPL1, APPL, DIP13A, KIAA1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
DP13A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.023577 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
IRS2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.028041 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
LAMB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.013276 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
MRIP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.056249 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
PLCG1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.016974 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P19174, Q2V575 | Gene names | PLCG1, PLC1 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-1) (Phospholipase C-gamma-1) (PLC-II) (PLC-148). | |||||
|
FAK2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | -0.000228 (rank : 93) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QVP9 | Gene names | Ptk2b, Fak2, Pyk2, Raftk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine kinase 2 beta (EC 2.7.10.2) (Focal adhesion kinase 2) (FADK 2) (Proline-rich tyrosine kinase 2) (Cell adhesion kinase beta) (CAK beta) (Calcium-dependent tyrosine kinase) (CADTK) (Related adhesion focal tyrosine kinase) (RAFTK). | |||||
|
PKHA1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.068728 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PLCG1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.021859 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62077, Q6P1G1 | Gene names | Plcg1, Plcg-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-1) (Phospholipase C-gamma-1) (PLC-II) (PLC-148). | |||||
|
AKA11_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.022741 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKA4, O75124, Q9NUK7 | Gene names | AKAP11, AKAP220, KIAA0629 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 11 (Protein kinase A-anchoring protein 11) (PRKA11) (A kinase anchor protein 220 kDa) (AKAP 220) (hAKAP220). | |||||
|
MRCKA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.004122 (rank : 90) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
NEB2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.017333 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
TRI14_MOUSE
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.012558 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVW3, Q6A0C3, Q762I6, Q9D3G8 | Gene names | Trim14, Kiaa0129, Pub | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 14 (PU.1-binding protein). | |||||
|
ARK72_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.017446 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43488, O75749, Q5TG63 | Gene names | AKR7A2, AFAR, AFAR1, AKR7 | |||
|
Domain Architecture |

|
|||||
| Description | Aflatoxin B1 aldehyde reductase member 2 (EC 1.-.-.-) (AFB1-AR 1) (Aldoketoreductase 7). | |||||
|
CNKR1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.047415 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q969H4, O95381 | Gene names | CNKSR1, CNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 1 (Connector enhancer of KSR1) (hCNK1) (Connector enhancer of KSR-like) (CNK homolog protein 1). | |||||
|
CTNA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.014617 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26232, Q4ZFW1, Q53R26, Q53R33, Q53T67, Q53T71, Q53TM8, Q7Z3Y0 | Gene names | CTNNA2, CAPR | |||
|
Domain Architecture |

|
|||||
| Description | Catenin alpha-2 (Alpha-catenin-related protein) (Alpha N-catenin). | |||||
|
CTNA2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.014626 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61301, Q61300, Q6AXD1 | Gene names | Ctnna2, Catna2 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin alpha-2 (Alpha-catenin-related protein) (Alpha N-catenin). | |||||
|
DMXL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.009986 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y485 | Gene names | DMXL1, XL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
PKHA4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.095333 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.025709 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
TTC7A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.013435 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULT0, Q6PIX4, Q8ND67, Q9BUS3 | Gene names | TTC7A, KIAA1140, TTC7 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7A (TPR repeat protein 7). | |||||
|
IFT81_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.022814 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
MRIP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.055975 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
PKHA4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.088553 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
ACHB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.004764 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17787, Q9UEH9 | Gene names | CHRNB2 | |||
|
Domain Architecture |
|
|||||
| Description | Neuronal acetylcholine receptor protein subunit beta-2 precursor. | |||||
|
ACHB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.004755 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ERK7 | Gene names | Chrnb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal acetylcholine receptor protein subunit beta-2 precursor. | |||||
|
ACOX3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.009015 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EPL9 | Gene names | Acox3 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 3, peroxisomal (EC 1.3.3.6) (Pristanoyl-CoA oxidase) (Branched-chain acyl-CoA oxidase) (BRCACox). | |||||
|
ARK72_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.015388 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CG76, Q3UPU2, Q8CG77, Q8JZQ8, Q9D157 | Gene names | Akr7a2, Afar, Akr7a5 | |||
|
Domain Architecture |
|
|||||
| Description | Aflatoxin B1 aldehyde reductase member 2 (EC 1.-.-.-). | |||||
|
CALR3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.009429 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D9Q6 | Gene names | Calr3, Crt2 | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin-3 precursor (Calreticulin-2). | |||||
|
K2C6B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.005661 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P04259, P48669 | Gene names | KRT6B, K6B | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6B (Cytokeratin-6B) (CK 6B) (K6b keratin). | |||||
|
K2C6E_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.005597 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P48668, Q7RTN9 | Gene names | KRT6E | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6E (Cytokeratin-6E) (CK 6E) (K6e keratin) (Keratin K6h). | |||||
|
NEB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.015863 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
PLEK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.077407 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
RHES_MOUSE
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.003279 (rank : 91) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P63032, Q9WVD3 | Gene names | Rasd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein Rhes (Ras homolog enriched in striatum). | |||||
|
SIRT1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.009194 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q923E4, Q9QXG8 | Gene names | Sirt1, Sir2l1 | |||
|
Domain Architecture |

|
|||||
| Description | NAD-dependent deacetylase sirtuin-1 (EC 3.5.1.-) (SIR2alpha) (mSIR2a) (Sir2) (SIR2-like protein 1). | |||||
|
SPTN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.031854 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
SPTN4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.024374 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
ARHGG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.006313 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VV41, Q86TF0, Q99434 | Gene names | ARHGEF16, NBR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
IFT52_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.011680 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y366, Q5H8Z0, Q9H1G3, Q9H1G4, Q9H1H2 | Gene names | IFT52, C20orf9, NGD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 52 homolog (Protein NGD5 homolog). | |||||
|
RAI14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.009291 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
AKAP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.007533 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
CAH4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.004127 (rank : 89) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64444 | Gene names | Ca4, Car4 | |||
|
Domain Architecture |

|
|||||
| Description | Carbonic anhydrase 4 precursor (EC 4.2.1.1) (Carbonic anhydrase IV) (Carbonate dehydratase IV) (CA-IV). | |||||
|
CLH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.006204 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P53675, Q14017, Q15808, Q15809 | Gene names | CLTCL1, CLH22, CLTCL, CLTD | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 2 (CLH-22). | |||||
|
RHES_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.002344 (rank : 92) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96D21, O95520, Q5THY8 | Gene names | RASD2, TEM2 | |||
|
Domain Architecture |
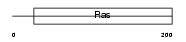
|
|||||
| Description | GTP-binding protein Rhes (Ras homolog enriched in striatum) (Tumor endothelial marker 2). | |||||
|
DAPP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.059848 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UN19, Q8TCK5, Q9UHF2 | Gene names | DAPP1, BAM32 | |||
|
Domain Architecture |
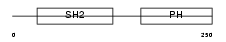
|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (hDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
DAPP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.059837 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QXT1, Q9R178 | Gene names | Dapp1, Bam32 | |||
|
Domain Architecture |
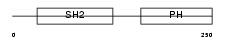
|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (mDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
GAB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.101432 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13480, Q4W5G2, Q6P1W2 | Gene names | GAB1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
IRS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.066127 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35568 | Gene names | IRS1 | |||
|
Domain Architecture |
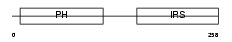
|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
IRS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.064455 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
DOC10_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q96BY6, O75178, Q9NW06, Q9NXI8 | Gene names | DOCK10, KIAA0694 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 10 (Protein zizimin 3). | |||||
|
DOCK9_MOUSE
|
||||||
| NC score | 0.978787 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BIK4, Q921Y6 | Gene names | Dock9, D14Wsu89e, Kiaa1058 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 9 (Cdc42 guanine nucleotide exchange factor zizimin 1). | |||||
|
DOCK9_HUMAN
|
||||||
| NC score | 0.977872 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BZ29, Q5JUD4, Q5JUD6, Q5T2Q1, Q5TAN8, Q9BZ25, Q9BZ26, Q9BZ27, Q9BZ28, Q9UPU4 | Gene names | DOCK9, KIAA1058 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 9 (Cdc42 guanine nucleotide exchange factor zizimin 1). | |||||
|
DOCK7_HUMAN
|
||||||
| NC score | 0.913205 (rank : 4) | θ value | 4.72459e-150 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96N67, Q6ZV32, Q8TB82, Q96NG6, Q9C092 | Gene names | DOCK7, KIAA1771 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dedicator of cytokinesis protein 7. | |||||
|
DOCK6_HUMAN
|
||||||
| NC score | 0.912800 (rank : 5) | θ value | 1.28961e-139 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96HP0, Q9P2F2 | Gene names | DOCK6, KIAA1395 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 6. | |||||
|
DOCK8_MOUSE
|
||||||
| NC score | 0.912780 (rank : 6) | θ value | 4.58675e-137 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C147, Q6PIS0, Q7TMQ5, Q8K105, Q9DBQ2 | Gene names | Dock8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dedicator of cytokinesis protein 8. | |||||
|
DOCK8_HUMAN
|
||||||
| NC score | 0.901250 (rank : 7) | θ value | 3.51195e-137 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NF50, Q3MV16, Q5JPJ1, Q8TEP1, Q8WUY2, Q9BYJ5, Q9H1Q2, Q9H1Q3, Q9H308, Q9H7P2 | Gene names | DOCK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dedicator of cytokinesis protein 8. | |||||
|
DOCK3_HUMAN
|
||||||
| NC score | 0.492703 (rank : 8) | θ value | 1.13037e-18 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IZD9, O15017 | Gene names | DOCK3, KIAA0299, MOCA | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 3 (Modifier of cell adhesion) (Presenilin-binding protein) (PBP protein). | |||||
|
DOCK3_MOUSE
|
||||||
| NC score | 0.492096 (rank : 9) | θ value | 3.28887e-18 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CIQ7 | Gene names | Dock3, Moca | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 3 (Modifier of cell adhesion) (Presenilin-binding protein) (PBP protein). | |||||
|
DOCK4_HUMAN
|
||||||
| NC score | 0.457602 (rank : 10) | θ value | 5.8054e-15 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK4_MOUSE
|
||||||
| NC score | 0.456588 (rank : 11) | θ value | 9.90251e-15 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK2_HUMAN
|
||||||
| NC score | 0.449803 (rank : 12) | θ value | 1.52774e-15 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92608, Q96AK7 | Gene names | DOCK2, KIAA0209 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 2. | |||||
|
DOCK2_MOUSE
|
||||||
| NC score | 0.448597 (rank : 13) | θ value | 6.85773e-16 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C3J5, Q99M79 | Gene names | Dock2 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 2 (Hch protein). | |||||
|
DOCK1_HUMAN
|
||||||
| NC score | 0.435049 (rank : 14) | θ value | 8.38298e-14 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14185 | Gene names | DOCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 1 (180 kDa protein downstream of CRK) (DOCK180). | |||||
|
GAB2_HUMAN
|
||||||
| NC score | 0.153334 (rank : 15) | θ value | 2.44474e-05 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UQC2, O60317 | Gene names | GAB2, KIAA0571 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (pp100). | |||||
|
GAB2_MOUSE
|
||||||
| NC score | 0.141770 (rank : 16) | θ value | 1.69304e-06 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
GAB1_MOUSE
|
||||||
| NC score | 0.121136 (rank : 17) | θ value | 0.0148317 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.115310 (rank : 18) | θ value | 7.1131e-05 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.111351 (rank : 19) | θ value | 0.0563607 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA5_HUMAN
|
||||||
| NC score | 0.105218 (rank : 20) | θ value | 0.0431538 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.103007 (rank : 21) | θ value | 0.00035302 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
GAB1_HUMAN
|
||||||
| NC score | 0.101432 (rank : 22) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13480, Q4W5G2, Q6P1W2 | Gene names | GAB1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
PKHA4_MOUSE
|
||||||
| NC score | 0.095333 (rank : 23) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PLEK_HUMAN
|
||||||
| NC score | 0.090924 (rank : 24) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
PKHA4_HUMAN
|
||||||
| NC score | 0.088553 (rank : 25) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PLEK_MOUSE
|
||||||
| NC score | 0.077407 (rank : 26) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
PLEK2_HUMAN
|
||||||
| NC score | 0.075688 (rank : 27) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NYT0, Q96JT0 | Gene names | PLEK2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin-2. | |||||
|
PLEK2_MOUSE
|
||||||
| NC score | 0.069338 (rank : 28) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WV52 | Gene names | Plek2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin-2. | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.069328 (rank : 29) | θ value | 0.163984 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA1_HUMAN
|
||||||
| NC score | 0.068728 (rank : 30) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
IRS1_HUMAN
|
||||||
| NC score | 0.066127 (rank : 31) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35568 | Gene names | IRS1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
IRS1_MOUSE
|
||||||
| NC score | 0.064455 (rank : 32) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.063153 (rank : 33) | θ value | 0.00869519 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PKHA1_MOUSE
|
||||||
| NC score | 0.060557 (rank : 34) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
DAPP1_HUMAN
|
||||||
| NC score | 0.059848 (rank : 35) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UN19, Q8TCK5, Q9UHF2 | Gene names | DAPP1, BAM32 | |||
|
Domain Architecture |

|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (hDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
DAPP1_MOUSE
|
||||||
| NC score | 0.059837 (rank : 36) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QXT1, Q9R178 | Gene names | Dapp1, Bam32 | |||
|
Domain Architecture |

|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (mDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
MRIP_MOUSE
|
||||||
| NC score | 0.056249 (rank : 37) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
MRIP_HUMAN
|
||||||
| NC score | 0.055975 (rank : 38) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.050195 (rank : 39) | θ value | 0.00509761 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CNKR1_HUMAN
|
||||||
| NC score | 0.047415 (rank : 40) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q969H4, O95381 | Gene names | CNKSR1, CNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 1 (Connector enhancer of KSR1) (hCNK1) (Connector enhancer of KSR-like) (CNK homolog protein 1). | |||||
|
NU107_HUMAN
|
||||||
| NC score | 0.044035 (rank : 41) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57740 | Gene names | NUP107 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup107 (Nucleoporin Nup107) (107 kDa nucleoporin). | |||||
|
NU107_MOUSE
|
||||||
| NC score | 0.038416 (rank : 42) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BH74, Q99KH5 | Gene names | Nup107 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup107 (Nucleoporin Nup107) (107 kDa nucleoporin). | |||||
|
SPTN2_HUMAN
|
||||||
| NC score | 0.031854 (rank : 43) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.031343 (rank : 44) | θ value | 0.00175202 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
IRS2_MOUSE
|
||||||
| NC score | 0.030170 (rank : 45) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
IRS2_HUMAN
|
||||||
| NC score | 0.028041 (rank : 46) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.026853 (rank : 47) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.025709 (rank : 48) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.025478 (rank : 49) | θ value | 0.0563607 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
IFT81_MOUSE
|
||||||
| NC score | 0.024687 (rank : 50) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
SPTN4_HUMAN
|
||||||
| NC score | 0.024374 (rank : 51) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
RPC1_HUMAN
|
||||||
| NC score | 0.023881 (rank : 52) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14802, Q8TCW5 | Gene names | POLR3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase III largest subunit (EC 2.7.7.6) (RPC155) (RPC1). | |||||
|
DP13A_HUMAN
|
||||||
| NC score | 0.023818 (rank : 53) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UKG1, Q9P2B9 | Gene names | APPL1, APPL, DIP13A, KIAA1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
DP13A_MOUSE
|
||||||
| NC score | 0.023577 (rank : 54) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
IFT81_HUMAN
|
||||||
| NC score | 0.022814 (rank : 55) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
AKA11_HUMAN
|
||||||
| NC score | 0.022741 (rank : 56) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKA4, O75124, Q9NUK7 | Gene names | AKAP11, AKAP220, KIAA0629 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 11 (Protein kinase A-anchoring protein 11) (PRKA11) (A kinase anchor protein 220 kDa) (AKAP 220) (hAKAP220). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.022247 (rank : 57) | θ value | 0.163984 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
PLCG1_MOUSE
|
||||||
| NC score | 0.021859 (rank : 58) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62077, Q6P1G1 | Gene names | Plcg1, Plcg-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-1) (Phospholipase C-gamma-1) (PLC-II) (PLC-148). | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | 0.021429 (rank : 59) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.019580 (rank : 60) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.019250 (rank : 61) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
ARK72_HUMAN
|
||||||
| NC score | 0.017446 (rank : 62) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43488, O75749, Q5TG63 | Gene names | AKR7A2, AFAR, AFAR1, AKR7 | |||
|
Domain Architecture |

|
|||||
| Description | Aflatoxin B1 aldehyde reductase member 2 (EC 1.-.-.-) (AFB1-AR 1) (Aldoketoreductase 7). | |||||
|
NEB2_MOUSE
|
||||||
| NC score | 0.017333 (rank : 63) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
PLCG1_HUMAN
|
||||||
| NC score | 0.016974 (rank : 64) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P19174, Q2V575 | Gene names | PLCG1, PLC1 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-1) (Phospholipase C-gamma-1) (PLC-II) (PLC-148). | |||||
|
NEB2_HUMAN
|
||||||
| NC score | 0.015863 (rank : 65) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
ARK72_MOUSE
|
||||||
| NC score | 0.015388 (rank : 66) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CG76, Q3UPU2, Q8CG77, Q8JZQ8, Q9D157 | Gene names | Akr7a2, Afar, Akr7a5 | |||
|
Domain Architecture |

|
|||||
| Description | Aflatoxin B1 aldehyde reductase member 2 (EC 1.-.-.-). | |||||
|
CTNA2_MOUSE
|
||||||
| NC score | 0.014626 (rank : 67) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61301, Q61300, Q6AXD1 | Gene names | Ctnna2, Catna2 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin alpha-2 (Alpha-catenin-related protein) (Alpha N-catenin). | |||||
|
CTNA2_HUMAN
|
||||||
| NC score | 0.014617 (rank : 68) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26232, Q4ZFW1, Q53R26, Q53R33, Q53T67, Q53T71, Q53TM8, Q7Z3Y0 | Gene names | CTNNA2, CAPR | |||
|
Domain Architecture |

|
|||||
| Description | Catenin alpha-2 (Alpha-catenin-related protein) (Alpha N-catenin). | |||||
|
TRI14_HUMAN
|
||||||
| NC score | 0.014299 (rank : 69) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14142, Q9BRD8, Q9C020 | Gene names | TRIM14, KIAA0129 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 14. | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.014002 (rank : 70) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
TTC7A_HUMAN
|
||||||
| NC score | 0.013435 (rank : 71) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULT0, Q6PIX4, Q8ND67, Q9BUS3 | Gene names | TTC7A, KIAA1140, TTC7 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7A (TPR repeat protein 7). | |||||
|
LAMB1_HUMAN
|
||||||
| NC score | 0.013276 (rank : 72) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
LAMB1_MOUSE
|
||||||
| NC score | 0.012900 (rank : 73) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
TRI14_MOUSE
|
||||||
| NC score | 0.012558 (rank : 74) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVW3, Q6A0C3, Q762I6, Q9D3G8 | Gene names | Trim14, Kiaa0129, Pub | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 14 (PU.1-binding protein). | |||||
|
IFT52_HUMAN
|
||||||
| NC score | 0.011680 (rank : 75) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y366, Q5H8Z0, Q9H1G3, Q9H1G4, Q9H1H2 | Gene names | IFT52, C20orf9, NGD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 52 homolog (Protein NGD5 homolog). | |||||
|
DMXL1_HUMAN
|
||||||
| NC score | 0.009986 (rank : 76) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y485 | Gene names | DMXL1, XL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
MPP2_MOUSE
|
||||||
| NC score | 0.009698 (rank : 77) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WV34 | Gene names | Mpp2, Dlgh2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 2 (Protein MPP2) (Dlgh2 protein). | |||||
|
CALR3_MOUSE
|
||||||
| NC score | 0.009429 (rank : 78) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D9Q6 | Gene names | Calr3, Crt2 | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin-3 precursor (Calreticulin-2). | |||||
|
RAI14_HUMAN
|
||||||
| NC score | 0.009291 (rank : 79) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
SIRT1_MOUSE
|
||||||
| NC score | 0.009194 (rank : 80) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q923E4, Q9QXG8 | Gene names | Sirt1, Sir2l1 | |||
|
Domain Architecture |

|
|||||
| Description | NAD-dependent deacetylase sirtuin-1 (EC 3.5.1.-) (SIR2alpha) (mSIR2a) (Sir2) (SIR2-like protein 1). | |||||
|
ACOX3_MOUSE
|
||||||
| NC score | 0.009015 (rank : 81) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EPL9 | Gene names | Acox3 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 3, peroxisomal (EC 1.3.3.6) (Pristanoyl-CoA oxidase) (Branched-chain acyl-CoA oxidase) (BRCACox). | |||||
|
AKAP1_HUMAN
|
||||||
| NC score | 0.007533 (rank : 82) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
ARHGG_HUMAN
|
||||||
| NC score | 0.006313 (rank : 83) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VV41, Q86TF0, Q99434 | Gene names | ARHGEF16, NBR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
CLH2_HUMAN
|
||||||
| NC score | 0.006204 (rank : 84) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P53675, Q14017, Q15808, Q15809 | Gene names | CLTCL1, CLH22, CLTCL, CLTD | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 2 (CLH-22). | |||||
|
K2C6B_HUMAN
|
||||||
| NC score | 0.005661 (rank : 85) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P04259, P48669 | Gene names | KRT6B, K6B | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6B (Cytokeratin-6B) (CK 6B) (K6b keratin). | |||||
|
K2C6E_HUMAN
|
||||||
| NC score | 0.005597 (rank : 86) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P48668, Q7RTN9 | Gene names | KRT6E | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6E (Cytokeratin-6E) (CK 6E) (K6e keratin) (Keratin K6h). | |||||
|
ACHB2_HUMAN
|
||||||
| NC score | 0.004764 (rank : 87) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17787, Q9UEH9 | Gene names | CHRNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal acetylcholine receptor protein subunit beta-2 precursor. | |||||
|
ACHB2_MOUSE
|
||||||
| NC score | 0.004755 (rank : 88) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ERK7 | Gene names | Chrnb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal acetylcholine receptor protein subunit beta-2 precursor. | |||||
|
CAH4_MOUSE
|
||||||
| NC score | 0.004127 (rank : 89) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64444 | Gene names | Ca4, Car4 | |||
|
Domain Architecture |

|
|||||
| Description | Carbonic anhydrase 4 precursor (EC 4.2.1.1) (Carbonic anhydrase IV) (Carbonate dehydratase IV) (CA-IV). | |||||
|
MRCKA_HUMAN
|
||||||
| NC score | 0.004122 (rank : 90) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
RHES_MOUSE
|
||||||
| NC score | 0.003279 (rank : 91) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P63032, Q9WVD3 | Gene names | Rasd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein Rhes (Ras homolog enriched in striatum). | |||||
|
RHES_HUMAN
|
||||||
| NC score | 0.002344 (rank : 92) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96D21, O95520, Q5THY8 | Gene names | RASD2, TEM2 | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein Rhes (Ras homolog enriched in striatum) (Tumor endothelial marker 2). | |||||
|
FAK2_MOUSE
|
||||||
| NC score | -0.000228 (rank : 93) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QVP9 | Gene names | Ptk2b, Fak2, Pyk2, Raftk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine kinase 2 beta (EC 2.7.10.2) (Focal adhesion kinase 2) (FADK 2) (Proline-rich tyrosine kinase 2) (Cell adhesion kinase beta) (CAK beta) (Calcium-dependent tyrosine kinase) (CADTK) (Related adhesion focal tyrosine kinase) (RAFTK). | |||||