Please be patient as the page loads
|
DOCK9_MOUSE
|
||||||
| SwissProt Accessions | Q8BIK4, Q921Y6 | Gene names | Dock9, D14Wsu89e, Kiaa1058 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 9 (Cdc42 guanine nucleotide exchange factor zizimin 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DOC10_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.978787 (rank : 3) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96BY6, O75178, Q9NW06, Q9NXI8 | Gene names | DOCK10, KIAA0694 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 10 (Protein zizimin 3). | |||||
|
DOCK6_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.923869 (rank : 4) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96HP0, Q9P2F2 | Gene names | DOCK6, KIAA1395 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 6. | |||||
|
DOCK9_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.996438 (rank : 2) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9BZ29, Q5JUD4, Q5JUD6, Q5T2Q1, Q5TAN8, Q9BZ25, Q9BZ26, Q9BZ27, Q9BZ28, Q9UPU4 | Gene names | DOCK9, KIAA1058 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 9 (Cdc42 guanine nucleotide exchange factor zizimin 1). | |||||
|
DOCK9_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8BIK4, Q921Y6 | Gene names | Dock9, D14Wsu89e, Kiaa1058 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 9 (Cdc42 guanine nucleotide exchange factor zizimin 1). | |||||
|
DOCK7_HUMAN
|
||||||
| θ value | 1.41594e-162 (rank : 5) | NC score | 0.917749 (rank : 5) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96N67, Q6ZV32, Q8TB82, Q96NG6, Q9C092 | Gene names | DOCK7, KIAA1771 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dedicator of cytokinesis protein 7. | |||||
|
DOCK8_HUMAN
|
||||||
| θ value | 1.12453e-151 (rank : 6) | NC score | 0.905137 (rank : 7) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NF50, Q3MV16, Q5JPJ1, Q8TEP1, Q8WUY2, Q9BYJ5, Q9H1Q2, Q9H1Q3, Q9H308, Q9H7P2 | Gene names | DOCK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dedicator of cytokinesis protein 8. | |||||
|
DOCK8_MOUSE
|
||||||
| θ value | 1.4755e-135 (rank : 7) | NC score | 0.915002 (rank : 6) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C147, Q6PIS0, Q7TMQ5, Q8K105, Q9DBQ2 | Gene names | Dock8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dedicator of cytokinesis protein 8. | |||||
|
DOCK3_HUMAN
|
||||||
| θ value | 3.63628e-17 (rank : 8) | NC score | 0.455938 (rank : 8) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IZD9, O15017 | Gene names | DOCK3, KIAA0299, MOCA | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 3 (Modifier of cell adhesion) (Presenilin-binding protein) (PBP protein). | |||||
|
DOCK3_MOUSE
|
||||||
| θ value | 8.10077e-17 (rank : 9) | NC score | 0.455303 (rank : 9) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CIQ7 | Gene names | Dock3, Moca | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 3 (Modifier of cell adhesion) (Presenilin-binding protein) (PBP protein). | |||||
|
DOCK4_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 10) | NC score | 0.418971 (rank : 10) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK4_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 11) | NC score | 0.417854 (rank : 11) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK2_MOUSE
|
||||||
| θ value | 6.64225e-11 (rank : 12) | NC score | 0.406678 (rank : 13) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C3J5, Q99M79 | Gene names | Dock2 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 2 (Hch protein). | |||||
|
DOCK1_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 13) | NC score | 0.394920 (rank : 14) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14185 | Gene names | DOCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 1 (180 kDa protein downstream of CRK) (DOCK180). | |||||
|
DOCK2_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 14) | NC score | 0.407731 (rank : 12) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92608, Q96AK7 | Gene names | DOCK2, KIAA0209 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 2. | |||||
|
DP13B_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 15) | NC score | 0.064964 (rank : 34) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEU8, Q8N4R7, Q9NVL2 | Gene names | DIP13B, APPL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 beta (Dip13 beta) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 2). | |||||
|
RHG25_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 16) | NC score | 0.115192 (rank : 18) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 17) | NC score | 0.116296 (rank : 16) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
SPTN2_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 18) | NC score | 0.045905 (rank : 39) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
RHG25_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 19) | NC score | 0.104855 (rank : 22) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
IRS1_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 20) | NC score | 0.095109 (rank : 24) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35568 | Gene names | IRS1 | |||
|
Domain Architecture |
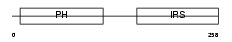
|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
IRS1_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 21) | NC score | 0.093657 (rank : 25) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
PKHA5_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 22) | NC score | 0.108455 (rank : 21) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
CENB1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 23) | NC score | 0.056388 (rank : 37) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
CENB2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 24) | NC score | 0.058771 (rank : 36) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
GAB2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 25) | NC score | 0.115411 (rank : 17) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
PKHA4_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 26) | NC score | 0.108952 (rank : 20) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
GAB1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 27) | NC score | 0.109079 (rank : 19) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
GAB2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 28) | NC score | 0.128184 (rank : 15) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UQC2, O60317 | Gene names | GAB2, KIAA0571 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (pp100). | |||||
|
PKHA4_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 29) | NC score | 0.104065 (rank : 23) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
DAPP1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 30) | NC score | 0.071159 (rank : 30) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UN19, Q8TCK5, Q9UHF2 | Gene names | DAPP1, BAM32 | |||
|
Domain Architecture |
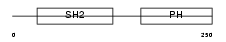
|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (hDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
DAPP1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 31) | NC score | 0.071369 (rank : 29) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QXT1, Q9R178 | Gene names | Dapp1, Bam32 | |||
|
Domain Architecture |
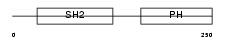
|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (mDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 0.163984 (rank : 32) | NC score | 0.018950 (rank : 53) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
GAB1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 33) | NC score | 0.091566 (rank : 26) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13480, Q4W5G2, Q6P1W2 | Gene names | GAB1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
UACA_HUMAN
|
||||||
| θ value | 0.163984 (rank : 34) | NC score | 0.016246 (rank : 59) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
OSBP1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.037739 (rank : 40) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P22059 | Gene names | OSBP, OSBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 1. | |||||
|
PLEK_HUMAN
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.087485 (rank : 27) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
HD_HUMAN
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.028630 (rank : 45) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.033902 (rank : 41) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.032915 (rank : 43) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
RDH12_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.011707 (rank : 67) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BYK4, Q91WA5, Q9D1Y4 | Gene names | Rdh12 | |||
|
Domain Architecture |
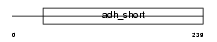
|
|||||
| Description | Retinol dehydrogenase 12 (EC 1.1.1.-). | |||||
|
PLEK_MOUSE
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.073690 (rank : 28) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
CHSTB_MOUSE
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.015922 (rank : 60) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JME2, Q9JJS2 | Gene names | Chst11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 11 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 1) (Chondroitin 4-sulfotransferase 1) (C4ST) (C4ST-1) (C4S-1). | |||||
|
K0226_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.025178 (rank : 47) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92622 | Gene names | KIAA0226 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0226. | |||||
|
SPTN4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.033078 (rank : 42) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
XPO4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.017025 (rank : 55) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ESJ0, Q6ZPJ4 | Gene names | Xpo4, Kiaa1721 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exportin-4 (Exp4). | |||||
|
NOC3L_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.026869 (rank : 46) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WTT2, Q9H5M6, Q9H9D8 | Gene names | NOC3L, AD24, C10orf117, FAD24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
PKHA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.066350 (rank : 33) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PLCG1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.020845 (rank : 48) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62077, Q6P1G1 | Gene names | Plcg1, Plcg-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-1) (Phospholipase C-gamma-1) (PLC-II) (PLC-148). | |||||
|
PLEK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.070202 (rank : 31) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NYT0, Q96JT0 | Gene names | PLEK2 | |||
|
Domain Architecture |
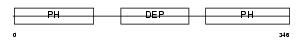
|
|||||
| Description | Pleckstrin-2. | |||||
|
DYR1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.003432 (rank : 77) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13627, O60769, Q92582, Q92810, Q9UNM5 | Gene names | DYRK1A, DYRK, MNB, MNBH | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity tyrosine-phosphorylation-regulated kinase 1A (EC 2.7.12.1) (Protein kinase minibrain homolog) (MNBH) (HP86) (Dual specificity YAK1-related kinase). | |||||
|
UBN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.029103 (rank : 44) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NPG3, Q13079, Q9P1P7 | Gene names | UBN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubinuclein (Ubiquitously expressed nuclear protein) (VT4). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.019402 (rank : 52) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.017320 (rank : 54) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.016707 (rank : 58) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MASTL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.000751 (rank : 79) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C0P0, Q5RJW0, Q6NXX9, Q8BVF3, Q9CZH9, Q9D9V0 | Gene names | Mastl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase-like (EC 2.7.11.1). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.012035 (rank : 66) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
CT112_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.019721 (rank : 51) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6DIB4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf112 homolog. | |||||
|
K0319_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.019903 (rank : 50) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VV43, Q9UJC8, Q9Y4G7 | Gene names | KIAA0319 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0319 precursor. | |||||
|
MASTL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.000434 (rank : 81) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 894 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96GX5, Q5T8D5, Q5T8D7, Q8NCD6, Q96SJ5 | Gene names | MASTL, THC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase-like (EC 2.7.11.1). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.008930 (rank : 69) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.006428 (rank : 75) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.008079 (rank : 72) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
CT011_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.016860 (rank : 56) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NWU2, Q8N5M5 | Gene names | C20orf11 | |||
|
Domain Architecture |
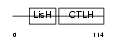
|
|||||
| Description | Protein C20orf11 (Two-hybrid associated protein 1 with RanBPM) (Twa1). | |||||
|
CT011_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.016776 (rank : 57) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D7M1 | Gene names | ||||
|
Domain Architecture |
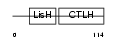
|
|||||
| Description | Protein C20orf11 homolog (Two-hybrid associated protein 1 with RanBPM) (Twa1). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.003269 (rank : 78) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CUL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.008203 (rank : 70) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13616, O60719, Q8IYW1 | Gene names | CUL1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CUL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.008203 (rank : 71) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WTX6, Q9WUI7 | Gene names | Cul1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
DMD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.020147 (rank : 49) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
HXA3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.000510 (rank : 80) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43365 | Gene names | HOXA3, HOX1E | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1E). | |||||
|
PLEK2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.061553 (rank : 35) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WV52 | Gene names | Plek2 | |||
|
Domain Architecture |
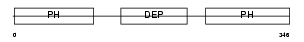
|
|||||
| Description | Pleckstrin-2. | |||||
|
RFC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.008005 (rank : 73) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
SDC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.012388 (rank : 65) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43407 | Gene names | Sdc2, Hspg1, Synd2 | |||
|
Domain Architecture |
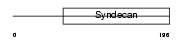
|
|||||
| Description | Syndecan-2 precursor (SYND2) (Fibroglycan) (Heparan sulfate proteoglycan core protein) (HSPG). | |||||
|
TTC3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.014287 (rank : 62) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P53804, O60767, P78476, P78477 | Gene names | TTC3, TPRD | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 3 (TPR repeat protein 3) (TPR repeat protein D). | |||||
|
ATM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.005612 (rank : 76) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13315, O15429, Q12758, Q16551, Q93007, Q9NP02, Q9UCX7 | Gene names | ATM | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated) (A-T, mutated). | |||||
|
BACH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.013260 (rank : 64) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00154, O43703, Q5JYL4, Q9UJM9, Q9Y539, Q9Y540 | Gene names | ACOT7, BACH | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic acyl coenzyme A thioester hydrolase (EC 3.1.2.2) (Long chain acyl-CoA thioester hydrolase) (CTE-II) (CTE-IIa) (Brain acyl-CoA hydrolase) (Acyl-CoA thioesterase 7). | |||||
|
FETUA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.009212 (rank : 68) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29699, O35634 | Gene names | Ahsg, Fetua | |||
|
Domain Architecture |
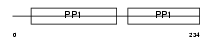
|
|||||
| Description | Alpha-2-HS-glycoprotein precursor (Fetuin-A) (Countertrypin). | |||||
|
FGD5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.013329 (rank : 63) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
LGP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.007994 (rank : 74) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
PLCG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.015250 (rank : 61) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P19174, Q2V575 | Gene names | PLCG1, PLC1 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-1) (Phospholipase C-gamma-1) (PLC-II) (PLC-148). | |||||
|
PKHA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.055316 (rank : 38) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.069024 (rank : 32) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
DOCK9_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8BIK4, Q921Y6 | Gene names | Dock9, D14Wsu89e, Kiaa1058 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 9 (Cdc42 guanine nucleotide exchange factor zizimin 1). | |||||
|
DOCK9_HUMAN
|
||||||
| NC score | 0.996438 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9BZ29, Q5JUD4, Q5JUD6, Q5T2Q1, Q5TAN8, Q9BZ25, Q9BZ26, Q9BZ27, Q9BZ28, Q9UPU4 | Gene names | DOCK9, KIAA1058 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 9 (Cdc42 guanine nucleotide exchange factor zizimin 1). | |||||
|
DOC10_HUMAN
|
||||||
| NC score | 0.978787 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96BY6, O75178, Q9NW06, Q9NXI8 | Gene names | DOCK10, KIAA0694 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 10 (Protein zizimin 3). | |||||
|
DOCK6_HUMAN
|
||||||
| NC score | 0.923869 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96HP0, Q9P2F2 | Gene names | DOCK6, KIAA1395 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 6. | |||||
|
DOCK7_HUMAN
|
||||||
| NC score | 0.917749 (rank : 5) | θ value | 1.41594e-162 (rank : 5) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96N67, Q6ZV32, Q8TB82, Q96NG6, Q9C092 | Gene names | DOCK7, KIAA1771 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dedicator of cytokinesis protein 7. | |||||
|
DOCK8_MOUSE
|
||||||
| NC score | 0.915002 (rank : 6) | θ value | 1.4755e-135 (rank : 7) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C147, Q6PIS0, Q7TMQ5, Q8K105, Q9DBQ2 | Gene names | Dock8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dedicator of cytokinesis protein 8. | |||||
|
DOCK8_HUMAN
|
||||||
| NC score | 0.905137 (rank : 7) | θ value | 1.12453e-151 (rank : 6) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NF50, Q3MV16, Q5JPJ1, Q8TEP1, Q8WUY2, Q9BYJ5, Q9H1Q2, Q9H1Q3, Q9H308, Q9H7P2 | Gene names | DOCK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dedicator of cytokinesis protein 8. | |||||
|
DOCK3_HUMAN
|
||||||
| NC score | 0.455938 (rank : 8) | θ value | 3.63628e-17 (rank : 8) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IZD9, O15017 | Gene names | DOCK3, KIAA0299, MOCA | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 3 (Modifier of cell adhesion) (Presenilin-binding protein) (PBP protein). | |||||
|
DOCK3_MOUSE
|
||||||
| NC score | 0.455303 (rank : 9) | θ value | 8.10077e-17 (rank : 9) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CIQ7 | Gene names | Dock3, Moca | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 3 (Modifier of cell adhesion) (Presenilin-binding protein) (PBP protein). | |||||
|
DOCK4_HUMAN
|
||||||
| NC score | 0.418971 (rank : 10) | θ value | 3.52202e-12 (rank : 10) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK4_MOUSE
|
||||||
| NC score | 0.417854 (rank : 11) | θ value | 4.59992e-12 (rank : 11) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK2_HUMAN
|
||||||
| NC score | 0.407731 (rank : 12) | θ value | 2.52405e-10 (rank : 14) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92608, Q96AK7 | Gene names | DOCK2, KIAA0209 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 2. | |||||
|
DOCK2_MOUSE
|
||||||
| NC score | 0.406678 (rank : 13) | θ value | 6.64225e-11 (rank : 12) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C3J5, Q99M79 | Gene names | Dock2 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 2 (Hch protein). | |||||
|
DOCK1_HUMAN
|
||||||
| NC score | 0.394920 (rank : 14) | θ value | 1.133e-10 (rank : 13) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14185 | Gene names | DOCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 1 (180 kDa protein downstream of CRK) (DOCK180). | |||||
|
GAB2_HUMAN
|
||||||
| NC score | 0.128184 (rank : 15) | θ value | 0.0563607 (rank : 28) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UQC2, O60317 | Gene names | GAB2, KIAA0571 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (pp100). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.116296 (rank : 16) | θ value | 0.000786445 (rank : 17) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
GAB2_MOUSE
|
||||||
| NC score | 0.115411 (rank : 17) | θ value | 0.0330416 (rank : 25) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.115192 (rank : 18) | θ value | 4.1701e-05 (rank : 16) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
GAB1_MOUSE
|
||||||
| NC score | 0.109079 (rank : 19) | θ value | 0.0563607 (rank : 27) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
PKHA4_MOUSE
|
||||||
| NC score | 0.108952 (rank : 20) | θ value | 0.0431538 (rank : 26) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PKHA5_HUMAN
|
||||||
| NC score | 0.108455 (rank : 21) | θ value | 0.00298849 (rank : 22) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.104855 (rank : 22) | θ value | 0.00102713 (rank : 19) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
PKHA4_HUMAN
|
||||||
| NC score | 0.104065 (rank : 23) | θ value | 0.0563607 (rank : 29) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
IRS1_HUMAN
|
||||||
| NC score | 0.095109 (rank : 24) | θ value | 0.00298849 (rank : 20) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35568 | Gene names | IRS1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
IRS1_MOUSE
|
||||||
| NC score | 0.093657 (rank : 25) | θ value | 0.00298849 (rank : 21) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
GAB1_HUMAN
|
||||||
| NC score | 0.091566 (rank : 26) | θ value | 0.163984 (rank : 33) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13480, Q4W5G2, Q6P1W2 | Gene names | GAB1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
PLEK_HUMAN
|
||||||
| NC score | 0.087485 (rank : 27) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
PLEK_MOUSE
|
||||||
| NC score | 0.073690 (rank : 28) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
DAPP1_MOUSE
|
||||||
| NC score | 0.071369 (rank : 29) | θ value | 0.125558 (rank : 31) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QXT1, Q9R178 | Gene names | Dapp1, Bam32 | |||
|
Domain Architecture |

|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (mDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
DAPP1_HUMAN
|
||||||
| NC score | 0.071159 (rank : 30) | θ value | 0.125558 (rank : 30) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UN19, Q8TCK5, Q9UHF2 | Gene names | DAPP1, BAM32 | |||
|
Domain Architecture |

|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (hDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
PLEK2_HUMAN
|
||||||
| NC score | 0.070202 (rank : 31) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NYT0, Q96JT0 | Gene names | PLEK2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin-2. | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.069024 (rank : 32) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA1_HUMAN
|
||||||
| NC score | 0.066350 (rank : 33) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
DP13B_HUMAN
|
||||||
| NC score | 0.064964 (rank : 34) | θ value | 4.1701e-05 (rank : 15) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEU8, Q8N4R7, Q9NVL2 | Gene names | DIP13B, APPL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 beta (Dip13 beta) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 2). | |||||
|
PLEK2_MOUSE
|
||||||
| NC score | 0.061553 (rank : 35) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WV52 | Gene names | Plek2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin-2. | |||||
|
CENB2_HUMAN
|
||||||
| NC score | 0.058771 (rank : 36) | θ value | 0.0193708 (rank : 24) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
CENB1_HUMAN
|
||||||
| NC score | 0.056388 (rank : 37) | θ value | 0.00665767 (rank : 23) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
PKHA1_MOUSE
|
||||||
| NC score | 0.055316 (rank : 38) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
SPTN2_HUMAN
|
||||||
| NC score | 0.045905 (rank : 39) | θ value | 0.000786445 (rank : 18) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
OSBP1_HUMAN
|
||||||
| NC score | 0.037739 (rank : 40) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P22059 | Gene names | OSBP, OSBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 1. | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.033902 (rank : 41) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTN4_HUMAN
|
||||||
| NC score | 0.033078 (rank : 42) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.032915 (rank : 43) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
UBN1_HUMAN
|
||||||
| NC score | 0.029103 (rank : 44) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NPG3, Q13079, Q9P1P7 | Gene names | UBN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubinuclein (Ubiquitously expressed nuclear protein) (VT4). | |||||
|
HD_HUMAN
|
||||||
| NC score | 0.028630 (rank : 45) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
NOC3L_HUMAN
|
||||||
| NC score | 0.026869 (rank : 46) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WTT2, Q9H5M6, Q9H9D8 | Gene names | NOC3L, AD24, C10orf117, FAD24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
K0226_HUMAN
|
||||||
| NC score | 0.025178 (rank : 47) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92622 | Gene names | KIAA0226 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0226. | |||||
|
PLCG1_MOUSE
|
||||||
| NC score | 0.020845 (rank : 48) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62077, Q6P1G1 | Gene names | Plcg1, Plcg-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-1) (Phospholipase C-gamma-1) (PLC-II) (PLC-148). | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.020147 (rank : 49) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
K0319_HUMAN
|
||||||
| NC score | 0.019903 (rank : 50) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VV43, Q9UJC8, Q9Y4G7 | Gene names | KIAA0319 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0319 precursor. | |||||
|
CT112_MOUSE
|
||||||
| NC score | 0.019721 (rank : 51) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6DIB4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf112 homolog. | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.019402 (rank : 52) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.018950 (rank : 53) | θ value | 0.163984 (rank : 32) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.017320 (rank : 54) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
XPO4_MOUSE
|
||||||
| NC score | 0.017025 (rank : 55) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ESJ0, Q6ZPJ4 | Gene names | Xpo4, Kiaa1721 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exportin-4 (Exp4). | |||||
|
CT011_HUMAN
|
||||||
| NC score | 0.016860 (rank : 56) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NWU2, Q8N5M5 | Gene names | C20orf11 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C20orf11 (Two-hybrid associated protein 1 with RanBPM) (Twa1). | |||||
|
CT011_MOUSE
|
||||||
| NC score | 0.016776 (rank : 57) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D7M1 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C20orf11 homolog (Two-hybrid associated protein 1 with RanBPM) (Twa1). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.016707 (rank : 58) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
UACA_HUMAN
|
||||||
| NC score | 0.016246 (rank : 59) | θ value | 0.163984 (rank : 34) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
CHSTB_MOUSE
|
||||||
| NC score | 0.015922 (rank : 60) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JME2, Q9JJS2 | Gene names | Chst11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 11 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 1) (Chondroitin 4-sulfotransferase 1) (C4ST) (C4ST-1) (C4S-1). | |||||
|
PLCG1_HUMAN
|
||||||
| NC score | 0.015250 (rank : 61) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P19174, Q2V575 | Gene names | PLCG1, PLC1 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-1) (Phospholipase C-gamma-1) (PLC-II) (PLC-148). | |||||
|
TTC3_HUMAN
|
||||||
| NC score | 0.014287 (rank : 62) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P53804, O60767, P78476, P78477 | Gene names | TTC3, TPRD | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 3 (TPR repeat protein 3) (TPR repeat protein D). | |||||
|
FGD5_HUMAN
|
||||||
| NC score | 0.013329 (rank : 63) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
BACH_HUMAN
|
||||||
| NC score | 0.013260 (rank : 64) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00154, O43703, Q5JYL4, Q9UJM9, Q9Y539, Q9Y540 | Gene names | ACOT7, BACH | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic acyl coenzyme A thioester hydrolase (EC 3.1.2.2) (Long chain acyl-CoA thioester hydrolase) (CTE-II) (CTE-IIa) (Brain acyl-CoA hydrolase) (Acyl-CoA thioesterase 7). | |||||
|
SDC2_MOUSE
|
||||||
| NC score | 0.012388 (rank : 65) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43407 | Gene names | Sdc2, Hspg1, Synd2 | |||
|
Domain Architecture |

|
|||||
| Description | Syndecan-2 precursor (SYND2) (Fibroglycan) (Heparan sulfate proteoglycan core protein) (HSPG). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.012035 (rank : 66) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
RDH12_MOUSE
|
||||||
| NC score | 0.011707 (rank : 67) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BYK4, Q91WA5, Q9D1Y4 | Gene names | Rdh12 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 12 (EC 1.1.1.-). | |||||
|
FETUA_MOUSE
|
||||||
| NC score | 0.009212 (rank : 68) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29699, O35634 | Gene names | Ahsg, Fetua | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2-HS-glycoprotein precursor (Fetuin-A) (Countertrypin). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.008930 (rank : 69) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
CUL1_HUMAN
|
||||||
| NC score | 0.008203 (rank : 70) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13616, O60719, Q8IYW1 | Gene names | CUL1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CUL1_MOUSE
|
||||||
| NC score | 0.008203 (rank : 71) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WTX6, Q9WUI7 | Gene names | Cul1 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-1 (CUL-1). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.008079 (rank : 72) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
RFC1_MOUSE
|
||||||
| NC score | 0.008005 (rank : 73) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
LGP2_HUMAN
|
||||||
| NC score | 0.007994 (rank : 74) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.006428 (rank : 75) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ATM_HUMAN
|
||||||
| NC score | 0.005612 (rank : 76) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13315, O15429, Q12758, Q16551, Q93007, Q9NP02, Q9UCX7 | Gene names | ATM | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated) (A-T, mutated). | |||||
|
DYR1A_HUMAN
|
||||||
| NC score | 0.003432 (rank : 77) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13627, O60769, Q92582, Q92810, Q9UNM5 | Gene names | DYRK1A, DYRK, MNB, MNBH | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity tyrosine-phosphorylation-regulated kinase 1A (EC 2.7.12.1) (Protein kinase minibrain homolog) (MNBH) (HP86) (Dual specificity YAK1-related kinase). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.003269 (rank : 78) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
MASTL_MOUSE
|
||||||
| NC score | 0.000751 (rank : 79) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C0P0, Q5RJW0, Q6NXX9, Q8BVF3, Q9CZH9, Q9D9V0 | Gene names | Mastl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase-like (EC 2.7.11.1). | |||||
|
HXA3_HUMAN
|
||||||
| NC score | 0.000510 (rank : 80) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43365 | Gene names | HOXA3, HOX1E | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1E). | |||||
|
MASTL_HUMAN
|
||||||
| NC score | 0.000434 (rank : 81) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 894 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96GX5, Q5T8D5, Q5T8D7, Q8NCD6, Q96SJ5 | Gene names | MASTL, THC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase-like (EC 2.7.11.1). | |||||