Please be patient as the page loads
|
DP13B_HUMAN
|
||||||
| SwissProt Accessions | Q8NEU8, Q8N4R7, Q9NVL2 | Gene names | DIP13B, APPL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 beta (Dip13 beta) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DP13A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.952539 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UKG1, Q9P2B9 | Gene names | APPL1, APPL, DIP13A, KIAA1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
DP13A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.957921 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
DP13B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8NEU8, Q8N4R7, Q9NVL2 | Gene names | DIP13B, APPL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 beta (Dip13 beta) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 2). | |||||
|
DP13B_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.986139 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K3G9, Q99LT7 | Gene names | Dip13b, Appl2, Dip3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 beta (Dip13 beta) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 2). | |||||
|
CENB2_HUMAN
|
||||||
| θ value | 4.15078e-21 (rank : 5) | NC score | 0.381255 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
CENB5_HUMAN
|
||||||
| θ value | 1.2077e-20 (rank : 6) | NC score | 0.369168 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
RHG26_HUMAN
|
||||||
| θ value | 9.26847e-13 (rank : 7) | NC score | 0.333335 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
OPHN1_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 8) | NC score | 0.337092 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
OPHN1_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 9) | NC score | 0.330630 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
CENB1_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 10) | NC score | 0.325650 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
DOCK9_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 11) | NC score | 0.064964 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BIK4, Q921Y6 | Gene names | Dock9, D14Wsu89e, Kiaa1058 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 9 (Cdc42 guanine nucleotide exchange factor zizimin 1). | |||||
|
DDEF2_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 12) | NC score | 0.252436 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
DDEF2_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 13) | NC score | 0.249308 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
DGKK_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 14) | NC score | 0.108263 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
DDFL1_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 15) | NC score | 0.185463 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TDY4, Q6P9F4, Q86UY1, Q9NXK2 | Gene names | DDEFL1, UPLC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1 (Protein up- regulated in liver cancer 1). | |||||
|
DDEF1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 16) | NC score | 0.234116 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
DDFL1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 17) | NC score | 0.166039 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5U464 | Gene names | Ddefl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1. | |||||
|
FGD4_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 18) | NC score | 0.058301 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
MTDC_HUMAN
|
||||||
| θ value | 0.125558 (rank : 19) | NC score | 0.080402 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13995, Q53G90, Q53GV5, Q53S36 | Gene names | MTHFD2, NMDMC | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial precursor [Includes: NAD-dependent methylenetetrahydrofolate dehydrogenase (EC 1.5.1.15); Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9)]. | |||||
|
MTDC_MOUSE
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.072197 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P18155, Q3TMN4 | Gene names | Mthfd2, Nmdmc | |||
|
Domain Architecture |
|
|||||
| Description | Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial precursor [Includes: NAD-dependent methylenetetrahydrofolate dehydrogenase (EC 1.5.1.15); Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9)]. | |||||
|
FGD4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.048296 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96M96, Q6ULS2, Q8TCP6 | Gene names | FGD4, FRABP, ZFYVE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein) (Zinc finger FYVE domain-containing protein 6). | |||||
|
DGKD_HUMAN
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.078231 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q16760, Q14158, Q6PK55, Q8NG53 | Gene names | DGKD, KIAA0145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase delta (EC 2.7.1.107) (Diglyceride kinase delta) (DGK-delta) (DAG kinase delta) (130 kDa diacylglycerol kinase). | |||||
|
JIP1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.080274 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
DGKH_HUMAN
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.076327 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86XP1, Q5VZW0, Q6PI56, Q86XP2, Q8N3N0, Q8N7J9 | Gene names | DGKH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase eta (EC 2.7.1.107) (Diglyceride kinase eta) (DGK-eta) (DAG kinase eta). | |||||
|
POF1B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.021871 (rank : 61) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 494 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WVV4, Q5H9E9, Q5H9F0, Q8NG12, Q9H5Y2, Q9H738, Q9H744 | Gene names | POF1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein POF1B (Premature ovarian failure protein 1B). | |||||
|
MRCKG_MOUSE
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.003580 (rank : 83) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1466 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80UW5 | Gene names | Cdc42bpg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
ZN292_HUMAN
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.021719 (rank : 62) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60281, Q9H8G3, Q9H8J4 | Gene names | ZNF292, KIAA0530 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 292. | |||||
|
CYH2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.040753 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99418, Q92958 | Gene names | PSCD2, ARNO | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-2 (ARF nucleotide-binding site opener) (ARNO protein) (ARF exchange factor). | |||||
|
CYH2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.040692 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P63034, O89099, P97695, Q3UP39 | Gene names | Pscd2, Sec7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-2 (CLM2) (SEC7 homolog B) (msec7-2). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.014720 (rank : 68) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
ITPR1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.021336 (rank : 63) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11881, P20943, Q99LG5 | Gene names | Itpr1, Insp3r, Pcd6, Pcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (Inositol 1,4,5-trisphosphate-binding protein P400) (Purkinje cell protein 1) (Protein PCD-6). | |||||
|
KIF1B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.011788 (rank : 74) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60333, Q96Q94, Q9BV80, Q9P280 | Gene names | KIF1B, KIAA0591, KIAA1448 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1B (Klp). | |||||
|
2ACA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.020174 (rank : 64) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06190, Q06189, Q9NPQ5 | Gene names | PPP2R3A, PPP2R3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A 72/130 kDa regulatory subunit B (PP2A, subunit B, B''-PR72/PR130) (PP2A, subunit B, B72/B130 isoforms) (PP2A, subunit B, PR72/PR130 isoforms) (PP2A, subunit B, R3 isoform). | |||||
|
NOTC3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.016283 (rank : 65) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.011588 (rank : 75) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
FGD1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.040615 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
KIF1B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.010958 (rank : 78) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60575, Q9R0B4, Q9WVE5, Q9Z119 | Gene names | Kif1b | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1B. | |||||
|
TPM1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.013169 (rank : 69) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P09493, P09494, P10469, Q86W64, Q96IK2, Q9UCY9 | Gene names | TPM1, TMSA | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
TPM1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.013118 (rank : 70) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P58771, P02558, P19354, P46902, P99034 | Gene names | Tpm1, Tpm-1, Tpma | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
FGD1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.036440 (rank : 60) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
JIP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.071766 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
JIP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.061328 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13387, Q96G62, Q99771, Q9NZ59, Q9UKQ4 | Gene names | MAPK8IP2, IB2, JIP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
JIP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.058811 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ERE9, Q9CXI4 | Gene names | Mapk8ip2, Ib2, Jip2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
KMHN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.011028 (rank : 77) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.011301 (rank : 76) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
NOTC3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.015103 (rank : 67) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.016079 (rank : 66) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.012936 (rank : 71) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
CYH3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.037251 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08967, Q8CI93 | Gene names | Pscd3, Grp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1) (CLM-3). | |||||
|
LAMA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.005043 (rank : 82) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.009189 (rank : 79) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
VDP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.011898 (rank : 73) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z1Z0, Q3T9L9, Q3TH58, Q3U1C7, Q3UMW6, Q91WE7, Q99JZ5 | Gene names | Vdp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
ANGP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.006896 (rank : 81) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15389 | Gene names | ANGPT1, KIAA0003 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 precursor (ANG-1). | |||||
|
ANGP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.012242 (rank : 72) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15123, Q9NRR7, Q9P2Y7 | Gene names | ANGPT2 | |||
|
Domain Architecture |
|
|||||
| Description | Angiopoietin-2 precursor (ANG-2). | |||||
|
BFSP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.003249 (rank : 84) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6NVD9, Q63832, Q6P5N4, Q8VDD6 | Gene names | Bfsp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phakinin (Beaded filament structural protein 2) (Lens fiber cell beaded filament protein CP 49) (CP49) (49 kDa cytoskeletal protein). | |||||
|
CAR11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.007291 (rank : 80) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BXL7, Q548H3 | Gene names | CARD11, CARMA1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 11 (CARD-containing MAGUK protein 3) (Carma 1). | |||||
|
CYH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.037176 (rank : 59) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15438, Q9P123, Q9P124 | Gene names | PSCD1, D17S811E | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (SEC7 homolog B2-1). | |||||
|
MRCKA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.002849 (rank : 85) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
ARFG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.093307 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N6T3, Q96KC4, Q96T02, Q9NSU3, Q9NVF6, Q9UIL0 | Gene names | ARFGAP1, ARF1GAP | |||
|
Domain Architecture |
|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
ARFG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.095537 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EPJ9, Q3TI52, Q8BMM6, Q8BMQ7 | Gene names | Arfgap1, Arf1gap | |||
|
Domain Architecture |
|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
ARFG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.089592 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NP61, Q9BSC6, Q9H9J0, Q9NT10, Q9NUP5, Q9Y4V3, Q9Y4V4 | Gene names | ARFGAP3, ARFGAP1 | |||
|
Domain Architecture |
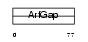
|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
ARFG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.088011 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D8S3, Q99KN8 | Gene names | Arfgap3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
CENA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.099627 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75689 | Gene names | CENTA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 1 (Putative MAPK-activating protein PM25). | |||||
|
CENA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.093016 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NPF8, Q8N4Q6, Q96SD5 | Gene names | CENTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-alpha 2. | |||||
|
CENA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.094428 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R2V5 | Gene names | Centa2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 2. | |||||
|
CEND1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.091077 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
CEND1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.092301 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
CEND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.095560 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
CEND3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.085962 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
CEND3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.084494 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
CENG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.077750 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99490, O00578, Q548E0, Q8IWU3 | Gene names | CENTG1, AGAP2, KIAA0167 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE) (GTP-binding and GTPase-activating protein 2) (GGAP2). | |||||
|
CENG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.078584 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
CENG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.072616 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPQ3, Q541S5, Q6P9D7, Q9NV93 | Gene names | CENTG2, AGAP1, KIAA1099 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1) (GTP-binding and GTPase-activating protein 1) (GGAP1). | |||||
|
CENG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.072691 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BXK8, Q3UHA0, Q6ZPX9, Q8BZG0 | Gene names | Centg2, Agap1, Kiaa1099 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1). | |||||
|
CENG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.076328 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96P47, Q59EN0, Q96RK3 | Gene names | CENTG3, AGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
CENG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.076647 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHH5, Q812F7 | Gene names | Centg3, Agap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
GIT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.061175 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y2X7, Q86SS0, Q9BRJ4 | Gene names | GIT1 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase- interactor 1) (GRK-interacting protein 1) (Cool-associated and tyrosine-phosphorylated protein 1) (Cat-1). | |||||
|
GIT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.061008 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q68FF6 | Gene names | Git1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase- interactor 1) (GRK-interacting protein 1). | |||||
|
GIT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.062689 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14161, Q86U59, Q96CI2, Q9BV91, Q9Y5V2 | Gene names | GIT2, KIAA0148 | |||
|
Domain Architecture |
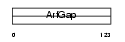
|
|||||
| Description | ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase- interactor 2) (GRK-interacting protein 2) (Cool-interacting tyrosine- phosphorylated protein 2) (CAT2) (CAT-2). | |||||
|
GIT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.062615 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JLQ2 | Gene names | Git2 | |||
|
Domain Architecture |
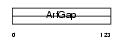
|
|||||
| Description | ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase- interactor 2) (Cool-interacting tyrosine-phosphorylated protein 2) (CAT2) (CAT-2). | |||||
|
NUPL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.050485 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P52594, Q15277, Q4VAS0, Q4VAS1, Q4VAS3, Q53QT8, Q53R11 | Gene names | HRB, RAB, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein) (Rev- interacting protein) (Rev/Rex activation domain-binding protein). | |||||
|
SMAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.094426 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
SMAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.096694 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
SMP1L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.094676 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WU79, Q5QPL2, Q96C93, Q9NST2, Q9UJL8 | Gene names | SMAP1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like. | |||||
|
SMP1L_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.094182 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TN29, Q3U798 | Gene names | Smap1l, Smap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like (Stromal membrane- associated protein 2). | |||||
|
DP13B_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8NEU8, Q8N4R7, Q9NVL2 | Gene names | DIP13B, APPL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 beta (Dip13 beta) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 2). | |||||
|
DP13B_MOUSE
|
||||||
| NC score | 0.986139 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K3G9, Q99LT7 | Gene names | Dip13b, Appl2, Dip3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 beta (Dip13 beta) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 2). | |||||
|
DP13A_MOUSE
|
||||||
| NC score | 0.957921 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
DP13A_HUMAN
|
||||||
| NC score | 0.952539 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UKG1, Q9P2B9 | Gene names | APPL1, APPL, DIP13A, KIAA1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
CENB2_HUMAN
|
||||||
| NC score | 0.381255 (rank : 5) | θ value | 4.15078e-21 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
CENB5_HUMAN
|
||||||
| NC score | 0.369168 (rank : 6) | θ value | 1.2077e-20 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
OPHN1_MOUSE
|
||||||
| NC score | 0.337092 (rank : 7) | θ value | 4.30538e-10 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
RHG26_HUMAN
|
||||||
| NC score | 0.333335 (rank : 8) | θ value | 9.26847e-13 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
OPHN1_HUMAN
|
||||||
| NC score | 0.330630 (rank : 9) | θ value | 8.11959e-09 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
CENB1_HUMAN
|
||||||
| NC score | 0.325650 (rank : 10) | θ value | 6.87365e-08 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
DDEF2_MOUSE
|
||||||
| NC score | 0.252436 (rank : 11) | θ value | 0.00020696 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
DDEF2_HUMAN
|
||||||
| NC score | 0.249308 (rank : 12) | θ value | 0.00102713 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
DDEF1_MOUSE
|
||||||
| NC score | 0.234116 (rank : 13) | θ value | 0.0113563 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
DDFL1_HUMAN
|
||||||
| NC score | 0.185463 (rank : 14) | θ value | 0.00298849 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TDY4, Q6P9F4, Q86UY1, Q9NXK2 | Gene names | DDEFL1, UPLC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1 (Protein up- regulated in liver cancer 1). | |||||
|
DDFL1_MOUSE
|
||||||
| NC score | 0.166039 (rank : 15) | θ value | 0.0563607 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5U464 | Gene names | Ddefl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1. | |||||
|
DGKK_HUMAN
|
||||||
| NC score | 0.108263 (rank : 16) | θ value | 0.00228821 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
CENA1_HUMAN
|
||||||
| NC score | 0.099627 (rank : 17) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75689 | Gene names | CENTA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 1 (Putative MAPK-activating protein PM25). | |||||
|
SMAP1_MOUSE
|
||||||
| NC score | 0.096694 (rank : 18) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
CEND2_HUMAN
|
||||||
| NC score | 0.095560 (rank : 19) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
ARFG1_MOUSE
|
||||||
| NC score | 0.095537 (rank : 20) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EPJ9, Q3TI52, Q8BMM6, Q8BMQ7 | Gene names | Arfgap1, Arf1gap | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
SMP1L_HUMAN
|
||||||
| NC score | 0.094676 (rank : 21) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WU79, Q5QPL2, Q96C93, Q9NST2, Q9UJL8 | Gene names | SMAP1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like. | |||||
|
CENA2_MOUSE
|
||||||
| NC score | 0.094428 (rank : 22) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R2V5 | Gene names | Centa2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 2. | |||||
|
SMAP1_HUMAN
|
||||||
| NC score | 0.094426 (rank : 23) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
SMP1L_MOUSE
|
||||||
| NC score | 0.094182 (rank : 24) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TN29, Q3U798 | Gene names | Smap1l, Smap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like (Stromal membrane- associated protein 2). | |||||
|
ARFG1_HUMAN
|
||||||
| NC score | 0.093307 (rank : 25) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N6T3, Q96KC4, Q96T02, Q9NSU3, Q9NVF6, Q9UIL0 | Gene names | ARFGAP1, ARF1GAP | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
CENA2_HUMAN
|
||||||
| NC score | 0.093016 (rank : 26) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NPF8, Q8N4Q6, Q96SD5 | Gene names | CENTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-alpha 2. | |||||
|
CEND1_MOUSE
|
||||||
| NC score | 0.092301 (rank : 27) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
CEND1_HUMAN
|
||||||
| NC score | 0.091077 (rank : 28) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
ARFG3_HUMAN
|
||||||
| NC score | 0.089592 (rank : 29) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NP61, Q9BSC6, Q9H9J0, Q9NT10, Q9NUP5, Q9Y4V3, Q9Y4V4 | Gene names | ARFGAP3, ARFGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
ARFG3_MOUSE
|
||||||
| NC score | 0.088011 (rank : 30) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D8S3, Q99KN8 | Gene names | Arfgap3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
CEND3_HUMAN
|
||||||
| NC score | 0.085962 (rank : 31) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
CEND3_MOUSE
|
||||||
| NC score | 0.084494 (rank : 32) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
MTDC_HUMAN
|
||||||
| NC score | 0.080402 (rank : 33) | θ value | 0.125558 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13995, Q53G90, Q53GV5, Q53S36 | Gene names | MTHFD2, NMDMC | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial precursor [Includes: NAD-dependent methylenetetrahydrofolate dehydrogenase (EC 1.5.1.15); Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9)]. | |||||
|
JIP1_HUMAN
|
||||||
| NC score | 0.080274 (rank : 34) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
CENG1_MOUSE
|
||||||
| NC score | 0.078584 (rank : 35) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
DGKD_HUMAN
|
||||||
| NC score | 0.078231 (rank : 36) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q16760, Q14158, Q6PK55, Q8NG53 | Gene names | DGKD, KIAA0145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase delta (EC 2.7.1.107) (Diglyceride kinase delta) (DGK-delta) (DAG kinase delta) (130 kDa diacylglycerol kinase). | |||||
|
CENG1_HUMAN
|
||||||
| NC score | 0.077750 (rank : 37) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99490, O00578, Q548E0, Q8IWU3 | Gene names | CENTG1, AGAP2, KIAA0167 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE) (GTP-binding and GTPase-activating protein 2) (GGAP2). | |||||
|
CENG3_MOUSE
|
||||||
| NC score | 0.076647 (rank : 38) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHH5, Q812F7 | Gene names | Centg3, Agap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
CENG3_HUMAN
|
||||||
| NC score | 0.076328 (rank : 39) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96P47, Q59EN0, Q96RK3 | Gene names | CENTG3, AGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
DGKH_HUMAN
|
||||||
| NC score | 0.076327 (rank : 40) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86XP1, Q5VZW0, Q6PI56, Q86XP2, Q8N3N0, Q8N7J9 | Gene names | DGKH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase eta (EC 2.7.1.107) (Diglyceride kinase eta) (DGK-eta) (DAG kinase eta). | |||||
|
CENG2_MOUSE
|
||||||
| NC score | 0.072691 (rank : 41) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BXK8, Q3UHA0, Q6ZPX9, Q8BZG0 | Gene names | Centg2, Agap1, Kiaa1099 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1). | |||||
|
CENG2_HUMAN
|
||||||
| NC score | 0.072616 (rank : 42) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPQ3, Q541S5, Q6P9D7, Q9NV93 | Gene names | CENTG2, AGAP1, KIAA1099 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1) (GTP-binding and GTPase-activating protein 1) (GGAP1). | |||||
|
MTDC_MOUSE
|
||||||
| NC score | 0.072197 (rank : 43) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P18155, Q3TMN4 | Gene names | Mthfd2, Nmdmc | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial precursor [Includes: NAD-dependent methylenetetrahydrofolate dehydrogenase (EC 1.5.1.15); Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9)]. | |||||
|
JIP1_MOUSE
|
||||||
| NC score | 0.071766 (rank : 44) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
DOCK9_MOUSE
|
||||||
| NC score | 0.064964 (rank : 45) | θ value | 4.1701e-05 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BIK4, Q921Y6 | Gene names | Dock9, D14Wsu89e, Kiaa1058 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 9 (Cdc42 guanine nucleotide exchange factor zizimin 1). | |||||
|
GIT2_HUMAN
|
||||||
| NC score | 0.062689 (rank : 46) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14161, Q86U59, Q96CI2, Q9BV91, Q9Y5V2 | Gene names | GIT2, KIAA0148 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase- interactor 2) (GRK-interacting protein 2) (Cool-interacting tyrosine- phosphorylated protein 2) (CAT2) (CAT-2). | |||||
|
GIT2_MOUSE
|
||||||
| NC score | 0.062615 (rank : 47) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JLQ2 | Gene names | Git2 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase- interactor 2) (Cool-interacting tyrosine-phosphorylated protein 2) (CAT2) (CAT-2). | |||||
|
JIP2_HUMAN
|
||||||
| NC score | 0.061328 (rank : 48) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13387, Q96G62, Q99771, Q9NZ59, Q9UKQ4 | Gene names | MAPK8IP2, IB2, JIP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
GIT1_HUMAN
|
||||||
| NC score | 0.061175 (rank : 49) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y2X7, Q86SS0, Q9BRJ4 | Gene names | GIT1 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase- interactor 1) (GRK-interacting protein 1) (Cool-associated and tyrosine-phosphorylated protein 1) (Cat-1). | |||||
|
GIT1_MOUSE
|
||||||
| NC score | 0.061008 (rank : 50) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q68FF6 | Gene names | Git1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase- interactor 1) (GRK-interacting protein 1). | |||||
|
JIP2_MOUSE
|
||||||
| NC score | 0.058811 (rank : 51) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ERE9, Q9CXI4 | Gene names | Mapk8ip2, Ib2, Jip2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
FGD4_MOUSE
|
||||||
| NC score | 0.058301 (rank : 52) | θ value | 0.0736092 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
NUPL_HUMAN
|
||||||
| NC score | 0.050485 (rank : 53) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P52594, Q15277, Q4VAS0, Q4VAS1, Q4VAS3, Q53QT8, Q53R11 | Gene names | HRB, RAB, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein) (Rev- interacting protein) (Rev/Rex activation domain-binding protein). | |||||
|
FGD4_HUMAN
|
||||||
| NC score | 0.048296 (rank : 54) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96M96, Q6ULS2, Q8TCP6 | Gene names | FGD4, FRABP, ZFYVE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein) (Zinc finger FYVE domain-containing protein 6). | |||||
|
CYH2_HUMAN
|
||||||
| NC score | 0.040753 (rank : 55) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99418, Q92958 | Gene names | PSCD2, ARNO | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-2 (ARF nucleotide-binding site opener) (ARNO protein) (ARF exchange factor). | |||||
|
CYH2_MOUSE
|
||||||
| NC score | 0.040692 (rank : 56) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P63034, O89099, P97695, Q3UP39 | Gene names | Pscd2, Sec7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-2 (CLM2) (SEC7 homolog B) (msec7-2). | |||||
|
FGD1_HUMAN
|
||||||
| NC score | 0.040615 (rank : 57) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
CYH3_MOUSE
|
||||||
| NC score | 0.037251 (rank : 58) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08967, Q8CI93 | Gene names | Pscd3, Grp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1) (CLM-3). | |||||
|
CYH1_HUMAN
|
||||||
| NC score | 0.037176 (rank : 59) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15438, Q9P123, Q9P124 | Gene names | PSCD1, D17S811E | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (SEC7 homolog B2-1). | |||||
|
FGD1_MOUSE
|
||||||
| NC score | 0.036440 (rank : 60) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
POF1B_HUMAN
|
||||||
| NC score | 0.021871 (rank : 61) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 494 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WVV4, Q5H9E9, Q5H9F0, Q8NG12, Q9H5Y2, Q9H738, Q9H744 | Gene names | POF1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein POF1B (Premature ovarian failure protein 1B). | |||||
|
ZN292_HUMAN
|
||||||
| NC score | 0.021719 (rank : 62) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60281, Q9H8G3, Q9H8J4 | Gene names | ZNF292, KIAA0530 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 292. | |||||
|
ITPR1_MOUSE
|
||||||
| NC score | 0.021336 (rank : 63) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11881, P20943, Q99LG5 | Gene names | Itpr1, Insp3r, Pcd6, Pcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (Inositol 1,4,5-trisphosphate-binding protein P400) (Purkinje cell protein 1) (Protein PCD-6). | |||||
|
2ACA_HUMAN
|
||||||
| NC score | 0.020174 (rank : 64) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06190, Q06189, Q9NPQ5 | Gene names | PPP2R3A, PPP2R3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A 72/130 kDa regulatory subunit B (PP2A, subunit B, B''-PR72/PR130) (PP2A, subunit B, B72/B130 isoforms) (PP2A, subunit B, PR72/PR130 isoforms) (PP2A, subunit B, R3 isoform). | |||||
|
NOTC3_HUMAN
|
||||||
| NC score | 0.016283 (rank : 65) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.016079 (rank : 66) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
NOTC3_MOUSE
|
||||||
| NC score | 0.015103 (rank : 67) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.014720 (rank : 68) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
TPM1_HUMAN
|
||||||
| NC score | 0.013169 (rank : 69) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P09493, P09494, P10469, Q86W64, Q96IK2, Q9UCY9 | Gene names | TPM1, TMSA | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
TPM1_MOUSE
|
||||||
| NC score | 0.013118 (rank : 70) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P58771, P02558, P19354, P46902, P99034 | Gene names | Tpm1, Tpm-1, Tpma | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.012936 (rank : 71) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
ANGP2_HUMAN
|
||||||
| NC score | 0.012242 (rank : 72) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15123, Q9NRR7, Q9P2Y7 | Gene names | ANGPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-2 precursor (ANG-2). | |||||
|
VDP_MOUSE
|
||||||
| NC score | 0.011898 (rank : 73) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z1Z0, Q3T9L9, Q3TH58, Q3U1C7, Q3UMW6, Q91WE7, Q99JZ5 | Gene names | Vdp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
KIF1B_HUMAN
|
||||||
| NC score | 0.011788 (rank : 74) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60333, Q96Q94, Q9BV80, Q9P280 | Gene names | KIF1B, KIAA0591, KIAA1448 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1B (Klp). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.011588 (rank : 75) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.011301 (rank : 76) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
KMHN1_HUMAN
|
||||||
| NC score | 0.011028 (rank : 77) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
KIF1B_MOUSE
|
||||||
| NC score | 0.010958 (rank : 78) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60575, Q9R0B4, Q9WVE5, Q9Z119 | Gene names | Kif1b | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1B. | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.009189 (rank : 79) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
CAR11_HUMAN
|
||||||
| NC score | 0.007291 (rank : 80) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BXL7, Q548H3 | Gene names | CARD11, CARMA1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 11 (CARD-containing MAGUK protein 3) (Carma 1). | |||||
|
ANGP1_HUMAN
|
||||||
| NC score | 0.006896 (rank : 81) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15389 | Gene names | ANGPT1, KIAA0003 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 precursor (ANG-1). | |||||
|
LAMA2_HUMAN
|
||||||
| NC score | 0.005043 (rank : 82) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
MRCKG_MOUSE
|
||||||
| NC score | 0.003580 (rank : 83) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1466 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80UW5 | Gene names | Cdc42bpg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
BFSP2_MOUSE
|
||||||
| NC score | 0.003249 (rank : 84) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6NVD9, Q63832, Q6P5N4, Q8VDD6 | Gene names | Bfsp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phakinin (Beaded filament structural protein 2) (Lens fiber cell beaded filament protein CP 49) (CP49) (49 kDa cytoskeletal protein). | |||||
|
MRCKA_HUMAN
|
||||||
| NC score | 0.002849 (rank : 85) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||