Please be patient as the page loads
|
NUPL_HUMAN
|
||||||
| SwissProt Accessions | P52594, Q15277, Q4VAS0, Q4VAS1, Q4VAS3, Q53QT8, Q53R11 | Gene names | HRB, RAB, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein) (Rev- interacting protein) (Rev/Rex activation domain-binding protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NUPL_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P52594, Q15277, Q4VAS0, Q4VAS1, Q4VAS3, Q53QT8, Q53R11 | Gene names | HRB, RAB, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein) (Rev- interacting protein) (Rev/Rex activation domain-binding protein). | |||||
|
NUPL_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.985829 (rank : 2) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K2K6, O70448, Q8BQL5, Q8CDK9 | Gene names | Hrb, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein homolog). | |||||
|
HRBL_MOUSE
|
||||||
| θ value | 5.00389e-67 (rank : 3) | NC score | 0.847500 (rank : 4) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
HRBL_HUMAN
|
||||||
| θ value | 2.48917e-58 (rank : 4) | NC score | 0.878232 (rank : 3) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
SMAP1_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 5) | NC score | 0.651969 (rank : 5) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
SMAP1_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 6) | NC score | 0.647527 (rank : 6) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
SMP1L_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 7) | NC score | 0.639209 (rank : 8) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WU79, Q5QPL2, Q96C93, Q9NST2, Q9UJL8 | Gene names | SMAP1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like. | |||||
|
SMP1L_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 8) | NC score | 0.640016 (rank : 7) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TN29, Q3U798 | Gene names | Smap1l, Smap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like (Stromal membrane- associated protein 2). | |||||
|
CENA1_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 9) | NC score | 0.614595 (rank : 9) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75689 | Gene names | CENTA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 1 (Putative MAPK-activating protein PM25). | |||||
|
ARFG1_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 10) | NC score | 0.589110 (rank : 10) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N6T3, Q96KC4, Q96T02, Q9NSU3, Q9NVF6, Q9UIL0 | Gene names | ARFGAP1, ARF1GAP | |||
|
Domain Architecture |
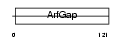
|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
CENG2_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 11) | NC score | 0.440333 (rank : 19) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BXK8, Q3UHA0, Q6ZPX9, Q8BZG0 | Gene names | Centg2, Agap1, Kiaa1099 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1). | |||||
|
CENG2_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 12) | NC score | 0.437143 (rank : 21) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UPQ3, Q541S5, Q6P9D7, Q9NV93 | Gene names | CENTG2, AGAP1, KIAA1099 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1) (GTP-binding and GTPase-activating protein 1) (GGAP1). | |||||
|
CENG3_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 13) | NC score | 0.443634 (rank : 17) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96P47, Q59EN0, Q96RK3 | Gene names | CENTG3, AGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
CENG3_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 14) | NC score | 0.444022 (rank : 16) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8VHH5, Q812F7 | Gene names | Centg3, Agap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
ARFG1_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 15) | NC score | 0.576834 (rank : 12) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9EPJ9, Q3TI52, Q8BMM6, Q8BMQ7 | Gene names | Arfgap1, Arf1gap | |||
|
Domain Architecture |
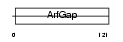
|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
CENA2_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 16) | NC score | 0.577606 (rank : 11) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8R2V5 | Gene names | Centa2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 2. | |||||
|
CENB2_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 17) | NC score | 0.397499 (rank : 24) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
CENB5_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 18) | NC score | 0.405287 (rank : 22) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
CENG1_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 19) | NC score | 0.443573 (rank : 18) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
CENG1_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 20) | NC score | 0.437593 (rank : 20) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99490, O00578, Q548E0, Q8IWU3 | Gene names | CENTG1, AGAP2, KIAA0167 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE) (GTP-binding and GTPase-activating protein 2) (GGAP2). | |||||
|
CENA2_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 21) | NC score | 0.566672 (rank : 13) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NPF8, Q8N4Q6, Q96SD5 | Gene names | CENTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-alpha 2. | |||||
|
ARFG3_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 22) | NC score | 0.526127 (rank : 15) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9D8S3, Q99KN8 | Gene names | Arfgap3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
ARFG3_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 23) | NC score | 0.528640 (rank : 14) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NP61, Q9BSC6, Q9H9J0, Q9NT10, Q9NUP5, Q9Y4V3, Q9Y4V4 | Gene names | ARFGAP3, ARFGAP1 | |||
|
Domain Architecture |
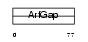
|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
CEND1_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 24) | NC score | 0.379255 (rank : 27) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
CEND3_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 25) | NC score | 0.372094 (rank : 31) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
CEND3_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 26) | NC score | 0.371707 (rank : 32) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
CEND1_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 27) | NC score | 0.375709 (rank : 30) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
GP112_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 28) | NC score | 0.054244 (rank : 52) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
DDEF1_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 29) | NC score | 0.386915 (rank : 26) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
DDEF2_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 30) | NC score | 0.377158 (rank : 29) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
CENB1_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 31) | NC score | 0.395319 (rank : 25) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
DDEF2_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 32) | NC score | 0.369260 (rank : 33) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
GIT2_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 33) | NC score | 0.319408 (rank : 35) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JLQ2 | Gene names | Git2 | |||
|
Domain Architecture |
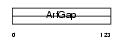
|
|||||
| Description | ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase- interactor 2) (Cool-interacting tyrosine-phosphorylated protein 2) (CAT2) (CAT-2). | |||||
|
CEND2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 34) | NC score | 0.357714 (rank : 34) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
DDFL1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 35) | NC score | 0.398982 (rank : 23) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TDY4, Q6P9F4, Q86UY1, Q9NXK2 | Gene names | DDEFL1, UPLC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1 (Protein up- regulated in liver cancer 1). | |||||
|
DDFL1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 36) | NC score | 0.377536 (rank : 28) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5U464 | Gene names | Ddefl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1. | |||||
|
GIT2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 37) | NC score | 0.309029 (rank : 36) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14161, Q86U59, Q96CI2, Q9BV91, Q9Y5V2 | Gene names | GIT2, KIAA0148 | |||
|
Domain Architecture |
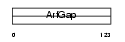
|
|||||
| Description | ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase- interactor 2) (GRK-interacting protein 2) (Cool-interacting tyrosine- phosphorylated protein 2) (CAT2) (CAT-2). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 38) | NC score | 0.032573 (rank : 63) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.033743 (rank : 61) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
PICAL_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.025774 (rank : 65) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13492, O60700, Q86XZ9 | Gene names | PICALM, CALM | |||
|
Domain Architecture |
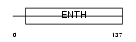
|
|||||
| Description | Phosphatidylinositol-binding clathrin assembly protein (Clathrin assembly lymphoid myeloid leukemia protein). | |||||
|
IF35_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.035584 (rank : 57) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
PDE4D_MOUSE
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.016742 (rank : 70) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01063, Q6TRH9, Q8C4Q7, Q8CG05 | Gene names | Pde4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3). | |||||
|
PDE4D_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.016225 (rank : 71) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08499, O43433, Q13549, Q13550, Q13551, Q7Z2L8, Q8IV84, Q8IVA9, Q8IVD2, Q8IVD3, Q9HCX7 | Gene names | PDE4D | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3) (PDE43). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.035929 (rank : 56) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.020780 (rank : 66) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
DAB2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.026760 (rank : 64) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
EYA4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.015386 (rank : 73) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95677, O95464, O95679, Q8IW39, Q9NTR7 | Gene names | EYA4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
MEGF9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.013695 (rank : 77) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
NU153_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.060644 (rank : 41) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
COE3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.015022 (rank : 74) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H4W6, Q5T6H9, Q9H4W5 | Gene names | EBF3, COE3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor COE3 (Early B-cell factor 3) (EBF-3) (Olf-1/EBF- like 2) (OE-2) (O/E-2). | |||||
|
TS1R2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.014543 (rank : 75) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q925I4, Q923J8 | Gene names | Tas1r2, Gpr71, T1r2, Tr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 2 precursor (G-protein coupled receptor 71) (Sweet taste receptor T1R2). | |||||
|
EP15_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.010835 (rank : 79) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
BUB1B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.015784 (rank : 72) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1S0 | Gene names | Bub1b, Mad3l | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta (EC 2.7.11.1) (MAD3/BUB1-related protein kinase) (Mitotic checkpoint kinase MAD3L). | |||||
|
CREM_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.019335 (rank : 67) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q03060, Q16114, Q16116, Q9NZB9 | Gene names | CREM | |||
|
Domain Architecture |
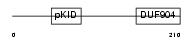
|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
EPN1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.035222 (rank : 58) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
RSMN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.034705 (rank : 59) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P63162, P14648, P17135 | Gene names | SNRPN, HCERN3, SMN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
RSMN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.034705 (rank : 60) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P63163, P14648, P17135 | Gene names | Snrpn, Smn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
GLI2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.001407 (rank : 82) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||
|
OSR1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | -0.001404 (rank : 84) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WVG7 | Gene names | Osr1, Odd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein odd-skipped-related 1. | |||||
|
TNKS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.040989 (rank : 55) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
CEP68_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.033619 (rank : 62) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q76N32, O60326, Q9BQ18, Q9UDM9 | Gene names | CEP68, KIAA0582 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
HIRA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.005426 (rank : 81) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54198, Q8IXN2 | Gene names | HIRA, DGCR1, HIR, TUPLE1 | |||
|
Domain Architecture |
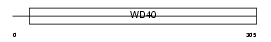
|
|||||
| Description | HIRA protein (TUP1-like enhancer of split protein 1). | |||||
|
P3H3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.012913 (rank : 78) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CG70, O88836 | Gene names | Leprel2, P3h3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 3 precursor (EC 1.14.11.7) (Leprecan-like protein 2) (Protein B). | |||||
|
TFR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.013900 (rank : 76) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62351, Q61560 | Gene names | Tfrc, Trfr | |||
|
Domain Architecture |

|
|||||
| Description | Transferrin receptor protein 1 (TfR1) (TR) (TfR) (Trfr) (CD71 antigen). | |||||
|
UBP5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.017628 (rank : 69) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.017630 (rank : 68) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
GLI3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | -0.000448 (rank : 83) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P10071, O75219, Q9UDT5, Q9UJ39 | Gene names | GLI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
LY75_MOUSE
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.010543 (rank : 80) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60767, Q8C7T3, Q91XL8, Q9QUZ6 | Gene names | Ly75, Cd205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (CD205 antigen). | |||||
|
ANR46_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.051785 (rank : 53) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86W74, Q6P9B7 | Gene names | ANKRD46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 46 (Ankyrin repeat small protein) (ANK-S). | |||||
|
CT086_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.055517 (rank : 50) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BZ19, Q4VXE6 | Gene names | C20orf86 | |||
|
Domain Architecture |

|
|||||
| Description | Putative uncharacterized protein C20orf86. | |||||
|
DP13A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.056741 (rank : 44) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UKG1, Q9P2B9 | Gene names | APPL1, APPL, DIP13A, KIAA1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
DP13A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.056599 (rank : 46) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
DP13B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.050485 (rank : 54) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NEU8, Q8N4R7, Q9NVL2 | Gene names | DIP13B, APPL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 beta (Dip13 beta) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 2). | |||||
|
DP13B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.054454 (rank : 51) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K3G9, Q99LT7 | Gene names | Dip13b, Appl2, Dip3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 beta (Dip13 beta) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 2). | |||||
|
ELN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.058422 (rank : 42) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15502, O15336, O15337, Q14233, Q14234, Q14235, Q14238, Q6P0L4, Q6ZWJ6, Q75MU5, Q7Z316, Q7Z3F5, Q9UMF5 | Gene names | ELN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elastin precursor (Tropoelastin). | |||||
|
ELN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.056522 (rank : 47) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54320 | Gene names | Eln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elastin precursor (Tropoelastin). | |||||
|
GIT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.272846 (rank : 37) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y2X7, Q86SS0, Q9BRJ4 | Gene names | GIT1 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase- interactor 1) (GRK-interacting protein 1) (Cool-associated and tyrosine-phosphorylated protein 1) (Cat-1). | |||||
|
GIT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.272199 (rank : 38) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q68FF6 | Gene names | Git1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase- interactor 1) (GRK-interacting protein 1). | |||||
|
OPHN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.061687 (rank : 40) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
OPHN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.066449 (rank : 39) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
PKHA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.056469 (rank : 48) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.055573 (rank : 49) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PYGO2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.056663 (rank : 45) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BRQ0, Q8WYZ4, Q96CY2 | Gene names | PYGO2 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 2. | |||||
|
RHG26_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.058406 (rank : 43) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
NUPL_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P52594, Q15277, Q4VAS0, Q4VAS1, Q4VAS3, Q53QT8, Q53R11 | Gene names | HRB, RAB, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein) (Rev- interacting protein) (Rev/Rex activation domain-binding protein). | |||||
|
NUPL_MOUSE
|
||||||
| NC score | 0.985829 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K2K6, O70448, Q8BQL5, Q8CDK9 | Gene names | Hrb, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein homolog). | |||||
|
HRBL_HUMAN
|
||||||
| NC score | 0.878232 (rank : 3) | θ value | 2.48917e-58 (rank : 4) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
HRBL_MOUSE
|
||||||
| NC score | 0.847500 (rank : 4) | θ value | 5.00389e-67 (rank : 3) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
SMAP1_HUMAN
|
||||||
| NC score | 0.651969 (rank : 5) | θ value | 3.52202e-12 (rank : 5) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
SMAP1_MOUSE
|
||||||
| NC score | 0.647527 (rank : 6) | θ value | 3.52202e-12 (rank : 6) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
SMP1L_MOUSE
|
||||||
| NC score | 0.640016 (rank : 7) | θ value | 3.89403e-11 (rank : 8) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TN29, Q3U798 | Gene names | Smap1l, Smap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like (Stromal membrane- associated protein 2). | |||||
|
SMP1L_HUMAN
|
||||||
| NC score | 0.639209 (rank : 8) | θ value | 3.89403e-11 (rank : 7) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WU79, Q5QPL2, Q96C93, Q9NST2, Q9UJL8 | Gene names | SMAP1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like. | |||||
|
CENA1_HUMAN
|
||||||
| NC score | 0.614595 (rank : 9) | θ value | 1.133e-10 (rank : 9) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75689 | Gene names | CENTA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 1 (Putative MAPK-activating protein PM25). | |||||
|
ARFG1_HUMAN
|
||||||
| NC score | 0.589110 (rank : 10) | θ value | 4.30538e-10 (rank : 10) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N6T3, Q96KC4, Q96T02, Q9NSU3, Q9NVF6, Q9UIL0 | Gene names | ARFGAP1, ARF1GAP | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
CENA2_MOUSE
|
||||||
| NC score | 0.577606 (rank : 11) | θ value | 1.69304e-06 (rank : 16) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8R2V5 | Gene names | Centa2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 2. | |||||
|
ARFG1_MOUSE
|
||||||
| NC score | 0.576834 (rank : 12) | θ value | 9.92553e-07 (rank : 15) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9EPJ9, Q3TI52, Q8BMM6, Q8BMQ7 | Gene names | Arfgap1, Arf1gap | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
CENA2_HUMAN
|
||||||
| NC score | 0.566672 (rank : 13) | θ value | 8.40245e-06 (rank : 21) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NPF8, Q8N4Q6, Q96SD5 | Gene names | CENTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-alpha 2. | |||||
|
ARFG3_HUMAN
|
||||||
| NC score | 0.528640 (rank : 14) | θ value | 4.1701e-05 (rank : 23) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NP61, Q9BSC6, Q9H9J0, Q9NT10, Q9NUP5, Q9Y4V3, Q9Y4V4 | Gene names | ARFGAP3, ARFGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
ARFG3_MOUSE
|
||||||
| NC score | 0.526127 (rank : 15) | θ value | 1.09739e-05 (rank : 22) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9D8S3, Q99KN8 | Gene names | Arfgap3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
CENG3_MOUSE
|
||||||
| NC score | 0.444022 (rank : 16) | θ value | 2.61198e-07 (rank : 14) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8VHH5, Q812F7 | Gene names | Centg3, Agap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
CENG3_HUMAN
|
||||||
| NC score | 0.443634 (rank : 17) | θ value | 2.61198e-07 (rank : 13) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96P47, Q59EN0, Q96RK3 | Gene names | CENTG3, AGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
CENG1_MOUSE
|
||||||
| NC score | 0.443573 (rank : 18) | θ value | 3.77169e-06 (rank : 19) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
CENG2_MOUSE
|
||||||
| NC score | 0.440333 (rank : 19) | θ value | 4.76016e-09 (rank : 11) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BXK8, Q3UHA0, Q6ZPX9, Q8BZG0 | Gene names | Centg2, Agap1, Kiaa1099 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1). | |||||
|
CENG1_HUMAN
|
||||||
| NC score | 0.437593 (rank : 20) | θ value | 4.92598e-06 (rank : 20) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99490, O00578, Q548E0, Q8IWU3 | Gene names | CENTG1, AGAP2, KIAA0167 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE) (GTP-binding and GTPase-activating protein 2) (GGAP2). | |||||
|
CENG2_HUMAN
|
||||||
| NC score | 0.437143 (rank : 21) | θ value | 1.38499e-08 (rank : 12) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UPQ3, Q541S5, Q6P9D7, Q9NV93 | Gene names | CENTG2, AGAP1, KIAA1099 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1) (GTP-binding and GTPase-activating protein 1) (GGAP1). | |||||
|
CENB5_HUMAN
|
||||||
| NC score | 0.405287 (rank : 22) | θ value | 3.77169e-06 (rank : 18) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
DDFL1_HUMAN
|
||||||
| NC score | 0.398982 (rank : 23) | θ value | 0.0113563 (rank : 35) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TDY4, Q6P9F4, Q86UY1, Q9NXK2 | Gene names | DDEFL1, UPLC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1 (Protein up- regulated in liver cancer 1). | |||||
|
CENB2_HUMAN
|
||||||
| NC score | 0.397499 (rank : 24) | θ value | 2.21117e-06 (rank : 17) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
CENB1_HUMAN
|
||||||
| NC score | 0.395319 (rank : 25) | θ value | 0.00175202 (rank : 31) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
DDEF1_MOUSE
|
||||||
| NC score | 0.386915 (rank : 26) | θ value | 0.000786445 (rank : 29) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
CEND1_MOUSE
|
||||||
| NC score | 0.379255 (rank : 27) | θ value | 4.1701e-05 (rank : 24) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
DDFL1_MOUSE
|
||||||
| NC score | 0.377536 (rank : 28) | θ value | 0.0148317 (rank : 36) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5U464 | Gene names | Ddefl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1. | |||||
|
DDEF2_HUMAN
|
||||||
| NC score | 0.377158 (rank : 29) | θ value | 0.00134147 (rank : 30) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
CEND1_HUMAN
|
||||||
| NC score | 0.375709 (rank : 30) | θ value | 0.00020696 (rank : 27) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
CEND3_MOUSE
|
||||||
| NC score | 0.372094 (rank : 31) | θ value | 9.29e-05 (rank : 25) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
CEND3_HUMAN
|
||||||
| NC score | 0.371707 (rank : 32) | θ value | 0.000121331 (rank : 26) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
DDEF2_MOUSE
|
||||||
| NC score | 0.369260 (rank : 33) | θ value | 0.00228821 (rank : 32) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
CEND2_HUMAN
|
||||||
| NC score | 0.357714 (rank : 34) | θ value | 0.0113563 (rank : 34) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
GIT2_MOUSE
|
||||||
| NC score | 0.319408 (rank : 35) | θ value | 0.00665767 (rank : 33) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JLQ2 | Gene names | Git2 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase- interactor 2) (Cool-interacting tyrosine-phosphorylated protein 2) (CAT2) (CAT-2). | |||||
|
GIT2_HUMAN
|
||||||
| NC score | 0.309029 (rank : 36) | θ value | 0.163984 (rank : 37) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14161, Q86U59, Q96CI2, Q9BV91, Q9Y5V2 | Gene names | GIT2, KIAA0148 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase- interactor 2) (GRK-interacting protein 2) (Cool-interacting tyrosine- phosphorylated protein 2) (CAT2) (CAT-2). | |||||
|
GIT1_HUMAN
|
||||||
| NC score | 0.272846 (rank : 37) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y2X7, Q86SS0, Q9BRJ4 | Gene names | GIT1 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase- interactor 1) (GRK-interacting protein 1) (Cool-associated and tyrosine-phosphorylated protein 1) (Cat-1). | |||||
|
GIT1_MOUSE
|
||||||
| NC score | 0.272199 (rank : 38) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q68FF6 | Gene names | Git1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase- interactor 1) (GRK-interacting protein 1). | |||||
|
OPHN1_MOUSE
|
||||||
| NC score | 0.066449 (rank : 39) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
OPHN1_HUMAN
|
||||||
| NC score | 0.061687 (rank : 40) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.060644 (rank : 41) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
ELN_HUMAN
|
||||||
| NC score | 0.058422 (rank : 42) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15502, O15336, O15337, Q14233, Q14234, Q14235, Q14238, Q6P0L4, Q6ZWJ6, Q75MU5, Q7Z316, Q7Z3F5, Q9UMF5 | Gene names | ELN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elastin precursor (Tropoelastin). | |||||
|
RHG26_HUMAN
|
||||||
| NC score | 0.058406 (rank : 43) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
DP13A_HUMAN
|
||||||
| NC score | 0.056741 (rank : 44) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UKG1, Q9P2B9 | Gene names | APPL1, APPL, DIP13A, KIAA1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
PYGO2_HUMAN
|
||||||
| NC score | 0.056663 (rank : 45) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BRQ0, Q8WYZ4, Q96CY2 | Gene names | PYGO2 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 2. | |||||
|
DP13A_MOUSE
|
||||||
| NC score | 0.056599 (rank : 46) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
ELN_MOUSE
|
||||||
| NC score | 0.056522 (rank : 47) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54320 | Gene names | Eln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elastin precursor (Tropoelastin). | |||||
|
PKHA1_HUMAN
|
||||||
| NC score | 0.056469 (rank : 48) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA1_MOUSE
|
||||||
| NC score | 0.055573 (rank : 49) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
CT086_HUMAN
|
||||||
| NC score | 0.055517 (rank : 50) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BZ19, Q4VXE6 | Gene names | C20orf86 | |||
|
Domain Architecture |

|
|||||
| Description | Putative uncharacterized protein C20orf86. | |||||
|
DP13B_MOUSE
|
||||||
| NC score | 0.054454 (rank : 51) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K3G9, Q99LT7 | Gene names | Dip13b, Appl2, Dip3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 beta (Dip13 beta) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 2). | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.054244 (rank : 52) | θ value | 0.000270298 (rank : 28) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
ANR46_HUMAN
|
||||||
| NC score | 0.051785 (rank : 53) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86W74, Q6P9B7 | Gene names | ANKRD46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 46 (Ankyrin repeat small protein) (ANK-S). | |||||
|
DP13B_HUMAN
|
||||||
| NC score | 0.050485 (rank : 54) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NEU8, Q8N4R7, Q9NVL2 | Gene names | DIP13B, APPL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 beta (Dip13 beta) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 2). | |||||
|
TNKS1_HUMAN
|
||||||
| NC score | 0.040989 (rank : 55) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.035929 (rank : 56) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
IF35_MOUSE
|
||||||
| NC score | 0.035584 (rank : 57) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
EPN1_MOUSE
|
||||||
| NC score | 0.035222 (rank : 58) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
RSMN_HUMAN
|
||||||
| NC score | 0.034705 (rank : 59) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P63162, P14648, P17135 | Gene names | SNRPN, HCERN3, SMN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
RSMN_MOUSE
|
||||||
| NC score | 0.034705 (rank : 60) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P63163, P14648, P17135 | Gene names | Snrpn, Smn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.033743 (rank : 61) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
CEP68_HUMAN
|
||||||
| NC score | 0.033619 (rank : 62) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q76N32, O60326, Q9BQ18, Q9UDM9 | Gene names | CEP68, KIAA0582 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.032573 (rank : 63) | θ value | 0.163984 (rank : 38) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
DAB2_HUMAN
|
||||||
| NC score | 0.026760 (rank : 64) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
PICAL_HUMAN
|
||||||
| NC score | 0.025774 (rank : 65) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13492, O60700, Q86XZ9 | Gene names | PICALM, CALM | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-binding clathrin assembly protein (Clathrin assembly lymphoid myeloid leukemia protein). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.020780 (rank : 66) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
CREM_HUMAN
|
||||||
| NC score | 0.019335 (rank : 67) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q03060, Q16114, Q16116, Q9NZB9 | Gene names | CREM | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
UBP5_MOUSE
|
||||||
| NC score | 0.017630 (rank : 68) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP5_HUMAN
|
||||||
| NC score | 0.017628 (rank : 69) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
PDE4D_MOUSE
|
||||||
| NC score | 0.016742 (rank : 70) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01063, Q6TRH9, Q8C4Q7, Q8CG05 | Gene names | Pde4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3). | |||||
|
PDE4D_HUMAN
|
||||||
| NC score | 0.016225 (rank : 71) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08499, O43433, Q13549, Q13550, Q13551, Q7Z2L8, Q8IV84, Q8IVA9, Q8IVD2, Q8IVD3, Q9HCX7 | Gene names | PDE4D | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3) (PDE43). | |||||
|
BUB1B_MOUSE
|
||||||
| NC score | 0.015784 (rank : 72) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1S0 | Gene names | Bub1b, Mad3l | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta (EC 2.7.11.1) (MAD3/BUB1-related protein kinase) (Mitotic checkpoint kinase MAD3L). | |||||
|
EYA4_HUMAN
|
||||||
| NC score | 0.015386 (rank : 73) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95677, O95464, O95679, Q8IW39, Q9NTR7 | Gene names | EYA4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
COE3_HUMAN
|
||||||
| NC score | 0.015022 (rank : 74) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H4W6, Q5T6H9, Q9H4W5 | Gene names | EBF3, COE3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor COE3 (Early B-cell factor 3) (EBF-3) (Olf-1/EBF- like 2) (OE-2) (O/E-2). | |||||
|
TS1R2_MOUSE
|
||||||
| NC score | 0.014543 (rank : 75) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q925I4, Q923J8 | Gene names | Tas1r2, Gpr71, T1r2, Tr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 2 precursor (G-protein coupled receptor 71) (Sweet taste receptor T1R2). | |||||
|
TFR1_MOUSE
|
||||||
| NC score | 0.013900 (rank : 76) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62351, Q61560 | Gene names | Tfrc, Trfr | |||
|
Domain Architecture |

|
|||||
| Description | Transferrin receptor protein 1 (TfR1) (TR) (TfR) (Trfr) (CD71 antigen). | |||||
|
MEGF9_HUMAN
|
||||||
| NC score | 0.013695 (rank : 77) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
P3H3_MOUSE
|
||||||
| NC score | 0.012913 (rank : 78) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CG70, O88836 | Gene names | Leprel2, P3h3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 3 precursor (EC 1.14.11.7) (Leprecan-like protein 2) (Protein B). | |||||
|
EP15_HUMAN
|
||||||
| NC score | 0.010835 (rank : 79) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
LY75_MOUSE
|
||||||
| NC score | 0.010543 (rank : 80) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60767, Q8C7T3, Q91XL8, Q9QUZ6 | Gene names | Ly75, Cd205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (CD205 antigen). | |||||
|
HIRA_HUMAN
|
||||||
| NC score | 0.005426 (rank : 81) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54198, Q8IXN2 | Gene names | HIRA, DGCR1, HIR, TUPLE1 | |||
|
Domain Architecture |

|
|||||
| Description | HIRA protein (TUP1-like enhancer of split protein 1). | |||||
|
GLI2_HUMAN
|
||||||
| NC score | 0.001407 (rank : 82) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||
|
GLI3_HUMAN
|
||||||
| NC score | -0.000448 (rank : 83) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P10071, O75219, Q9UDT5, Q9UJ39 | Gene names | GLI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
OSR1_MOUSE
|
||||||
| NC score | -0.001404 (rank : 84) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WVG7 | Gene names | Osr1, Odd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein odd-skipped-related 1. | |||||