Please be patient as the page loads
|
P3H3_MOUSE
|
||||||
| SwissProt Accessions | Q8CG70, O88836 | Gene names | Leprel2, P3h3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 3 precursor (EC 1.14.11.7) (Leprecan-like protein 2) (Protein B). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
P3H3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.996514 (rank : 2) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IVL6, Q13512, Q15740, Q66K32, Q6NX61, Q7L2T1 | Gene names | LEPREL2, P3H3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 3 precursor (EC 1.14.11.7) (Leprecan-like protein 2) (Protein B). | |||||
|
P3H3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CG70, O88836 | Gene names | Leprel2, P3h3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 3 precursor (EC 1.14.11.7) (Leprecan-like protein 2) (Protein B). | |||||
|
P3H1_HUMAN
|
||||||
| θ value | 1.09172e-138 (rank : 3) | NC score | 0.968213 (rank : 3) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q32P28, Q7KZR4, Q96BR8, Q96SK8, Q96SL5, Q96SN3, Q9H6K3, Q9HC86, Q9HC87 | Gene names | LEPRE1, GROS1, P3H1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 1 precursor (EC 1.14.11.7) (Leucine- and proline- enriched proteoglycan 1) (Leprecan-1) (Growth suppressor 1). | |||||
|
P3H1_MOUSE
|
||||||
| θ value | 1.20704e-137 (rank : 4) | NC score | 0.967732 (rank : 4) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3V1T4, Q3TWX8, Q8BSV2, Q8CFL3, Q9CWK5, Q9QZT6, Q9QZT7 | Gene names | Lepre1, Gros1, P3h1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 1 precursor (EC 1.14.11.7) (Leucine- and proline- enriched proteoglycan 1) (Leprecan-1) (Growth suppressor 1). | |||||
|
P3H2_HUMAN
|
||||||
| θ value | 1.6882e-131 (rank : 5) | NC score | 0.967055 (rank : 5) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IVL5, Q9NVI2 | Gene names | LEPREL1, MLAT4, P3H2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 2 precursor (EC 1.14.11.7) (Leprecan-like protein 1) (Myxoid liposarcoma-associated protein 4). | |||||
|
P3H2_MOUSE
|
||||||
| θ value | 1.17183e-124 (rank : 6) | NC score | 0.964763 (rank : 6) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CG71, Q8C673 | Gene names | Leprel1, P3h2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 2 precursor (EC 1.14.11.7) (Leprecan-like protein 1). | |||||
|
CRTAP_HUMAN
|
||||||
| θ value | 5.06226e-27 (rank : 7) | NC score | 0.816025 (rank : 7) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75718 | Gene names | CRTAP, CASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cartilage-associated protein precursor. | |||||
|
CRTAP_MOUSE
|
||||||
| θ value | 3.62785e-25 (rank : 8) | NC score | 0.806699 (rank : 8) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CYD3, O88698 | Gene names | Crtap, Casp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cartilage-associated protein precursor. | |||||
|
NO55_HUMAN
|
||||||
| θ value | 4.15078e-21 (rank : 9) | NC score | 0.793950 (rank : 9) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92791, Q9H4F6 | Gene names | SC65, NOL55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar autoantigen No55. | |||||
|
SKIV2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 10) | NC score | 0.052444 (rank : 11) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15477, O15005, Q12902, Q15476 | Gene names | SKIV2L, DDX13, SKI2W, SKIV2, W | |||
|
Domain Architecture |

|
|||||
| Description | Helicase SKI2W (EC 3.6.1.-) (Helicase-like protein) (HLP). | |||||
|
APEH_MOUSE
|
||||||
| θ value | 1.81305 (rank : 11) | NC score | 0.020509 (rank : 15) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R146, Q8K029, Q8R0M9 | Gene names | Apeh | |||
|
Domain Architecture |

|
|||||
| Description | Acylamino-acid-releasing enzyme (EC 3.4.19.1) (AARE) (Acyl-peptide hydrolase) (APH) (Acylaminoacyl-peptidase). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 12) | NC score | 0.001719 (rank : 28) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
FASTK_MOUSE
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.019279 (rank : 16) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JIX9 | Gene names | Fastk | |||
|
Domain Architecture |

|
|||||
| Description | Fas-activated serine/threonine kinase (EC 2.7.11.8) (FAST kinase). | |||||
|
KCNB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.005644 (rank : 22) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.001131 (rank : 29) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
I17RB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 16) | NC score | 0.013560 (rank : 17) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NRM6, Q9BPZ0, Q9NRL4, Q9NRM5 | Gene names | IL17RB, EVI27, IL17BR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-17 receptor B precursor (IL-17 receptor B) (IL-17RB) (Interleukin-17B receptor) (IL-17B receptor) (IL-17 receptor homolog 1) (IL-17Rh1) (IL17Rh1) (Cytokine receptor CRL4). | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.003875 (rank : 23) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
CP17A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.007631 (rank : 20) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P05093, Q5TZV7 | Gene names | CYP17A1, CYP17, S17AH | |||
|
Domain Architecture |
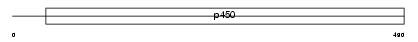
|
|||||
| Description | Cytochrome P450 17A1 (EC 1.14.99.9) (CYPXVII) (P450-C17) (P450c17) (Steroid 17-alpha-monooxygenase) (Steroid 17-alpha-hydroxylase/17,20 lyase). | |||||
|
EGLN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.025779 (rank : 14) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H6Z9 | Gene names | EGLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 3 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 3) (HIF-prolyl hydroxylase 3) (HIF-PH3) (HPH-1) (Prolyl hydroxylase domain-containing protein 3) (PHD3). | |||||
|
EGLN3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 20) | NC score | 0.025789 (rank : 13) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91UZ4, Q3TCG8, Q8C8M6, Q8CCA8, Q8R5C7 | Gene names | Egln3 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 3 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 3) (HIF-prolyl hydroxylase 3) (HIF-PH3) (SM-20). | |||||
|
FOXC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.003167 (rank : 27) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99958 | Gene names | FOXC2, FKHL14, MFH1 | |||
|
Domain Architecture |
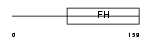
|
|||||
| Description | Forkhead box protein C2 (Forkhead-related protein FKHL14) (Mesenchyme fork head protein 1) (MFH-1 protein) (Transcription factor FKH-14). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.003812 (rank : 24) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
NUPL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.012913 (rank : 18) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52594, Q15277, Q4VAS0, Q4VAS1, Q4VAS3, Q53QT8, Q53R11 | Gene names | HRB, RAB, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein) (Rev- interacting protein) (Rev/Rex activation domain-binding protein). | |||||
|
NUPL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.012885 (rank : 19) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2K6, O70448, Q8BQL5, Q8CDK9 | Gene names | Hrb, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein homolog). | |||||
|
OXLA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.025821 (rank : 12) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O09046, Q3T9N9, Q3U7S6, Q3V0K2, Q6Y632, Q6YBV6, Q6YDI8, Q8R2G8, Q9CXK7 | Gene names | Il4i1, Fig1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (mFIG1). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.003274 (rank : 25) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
INADL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.003198 (rank : 26) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
SL9A2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.005784 (rank : 21) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBY0 | Gene names | SLC9A2, NHE2 | |||
|
Domain Architecture |
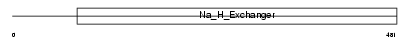
|
|||||
| Description | Sodium/hydrogen exchanger 2 (Na(+)/H(+) exchanger 2) (NHE-2) (Solute carrier family 9 member 2). | |||||
|
ZN287_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | -0.001943 (rank : 30) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBT7 | Gene names | ZNF287 | |||
|
Domain Architecture |
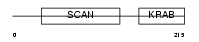
|
|||||
| Description | Zinc finger protein 287. | |||||
|
P4HA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.062291 (rank : 10) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60716, Q8VBU4 | Gene names | P4ha2 | |||
|
Domain Architecture |

|
|||||
| Description | Prolyl 4-hydroxylase alpha-2 subunit precursor (EC 1.14.11.2) (4-PH alpha-2) (Procollagen-proline,2-oxoglutarate-4-dioxygenase alpha-2 subunit). | |||||
|
P3H3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CG70, O88836 | Gene names | Leprel2, P3h3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 3 precursor (EC 1.14.11.7) (Leprecan-like protein 2) (Protein B). | |||||
|
P3H3_HUMAN
|
||||||
| NC score | 0.996514 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IVL6, Q13512, Q15740, Q66K32, Q6NX61, Q7L2T1 | Gene names | LEPREL2, P3H3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 3 precursor (EC 1.14.11.7) (Leprecan-like protein 2) (Protein B). | |||||
|
P3H1_HUMAN
|
||||||
| NC score | 0.968213 (rank : 3) | θ value | 1.09172e-138 (rank : 3) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q32P28, Q7KZR4, Q96BR8, Q96SK8, Q96SL5, Q96SN3, Q9H6K3, Q9HC86, Q9HC87 | Gene names | LEPRE1, GROS1, P3H1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 1 precursor (EC 1.14.11.7) (Leucine- and proline- enriched proteoglycan 1) (Leprecan-1) (Growth suppressor 1). | |||||
|
P3H1_MOUSE
|
||||||
| NC score | 0.967732 (rank : 4) | θ value | 1.20704e-137 (rank : 4) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3V1T4, Q3TWX8, Q8BSV2, Q8CFL3, Q9CWK5, Q9QZT6, Q9QZT7 | Gene names | Lepre1, Gros1, P3h1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 1 precursor (EC 1.14.11.7) (Leucine- and proline- enriched proteoglycan 1) (Leprecan-1) (Growth suppressor 1). | |||||
|
P3H2_HUMAN
|
||||||
| NC score | 0.967055 (rank : 5) | θ value | 1.6882e-131 (rank : 5) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IVL5, Q9NVI2 | Gene names | LEPREL1, MLAT4, P3H2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 2 precursor (EC 1.14.11.7) (Leprecan-like protein 1) (Myxoid liposarcoma-associated protein 4). | |||||
|
P3H2_MOUSE
|
||||||
| NC score | 0.964763 (rank : 6) | θ value | 1.17183e-124 (rank : 6) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CG71, Q8C673 | Gene names | Leprel1, P3h2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 2 precursor (EC 1.14.11.7) (Leprecan-like protein 1). | |||||
|
CRTAP_HUMAN
|
||||||
| NC score | 0.816025 (rank : 7) | θ value | 5.06226e-27 (rank : 7) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75718 | Gene names | CRTAP, CASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cartilage-associated protein precursor. | |||||
|
CRTAP_MOUSE
|
||||||
| NC score | 0.806699 (rank : 8) | θ value | 3.62785e-25 (rank : 8) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CYD3, O88698 | Gene names | Crtap, Casp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cartilage-associated protein precursor. | |||||
|
NO55_HUMAN
|
||||||
| NC score | 0.793950 (rank : 9) | θ value | 4.15078e-21 (rank : 9) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92791, Q9H4F6 | Gene names | SC65, NOL55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar autoantigen No55. | |||||
|
P4HA2_MOUSE
|
||||||
| NC score | 0.062291 (rank : 10) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60716, Q8VBU4 | Gene names | P4ha2 | |||
|
Domain Architecture |

|
|||||
| Description | Prolyl 4-hydroxylase alpha-2 subunit precursor (EC 1.14.11.2) (4-PH alpha-2) (Procollagen-proline,2-oxoglutarate-4-dioxygenase alpha-2 subunit). | |||||
|
SKIV2_HUMAN
|
||||||
| NC score | 0.052444 (rank : 11) | θ value | 0.0113563 (rank : 10) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15477, O15005, Q12902, Q15476 | Gene names | SKIV2L, DDX13, SKI2W, SKIV2, W | |||
|
Domain Architecture |

|
|||||
| Description | Helicase SKI2W (EC 3.6.1.-) (Helicase-like protein) (HLP). | |||||
|
OXLA_MOUSE
|
||||||
| NC score | 0.025821 (rank : 12) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O09046, Q3T9N9, Q3U7S6, Q3V0K2, Q6Y632, Q6YBV6, Q6YDI8, Q8R2G8, Q9CXK7 | Gene names | Il4i1, Fig1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (mFIG1). | |||||
|
EGLN3_MOUSE
|
||||||
| NC score | 0.025789 (rank : 13) | θ value | 6.88961 (rank : 20) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91UZ4, Q3TCG8, Q8C8M6, Q8CCA8, Q8R5C7 | Gene names | Egln3 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 3 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 3) (HIF-prolyl hydroxylase 3) (HIF-PH3) (SM-20). | |||||
|
EGLN3_HUMAN
|
||||||
| NC score | 0.025779 (rank : 14) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H6Z9 | Gene names | EGLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 3 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 3) (HIF-prolyl hydroxylase 3) (HIF-PH3) (HPH-1) (Prolyl hydroxylase domain-containing protein 3) (PHD3). | |||||
|
APEH_MOUSE
|
||||||
| NC score | 0.020509 (rank : 15) | θ value | 1.81305 (rank : 11) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R146, Q8K029, Q8R0M9 | Gene names | Apeh | |||
|
Domain Architecture |

|
|||||
| Description | Acylamino-acid-releasing enzyme (EC 3.4.19.1) (AARE) (Acyl-peptide hydrolase) (APH) (Acylaminoacyl-peptidase). | |||||
|
FASTK_MOUSE
|
||||||
| NC score | 0.019279 (rank : 16) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JIX9 | Gene names | Fastk | |||
|
Domain Architecture |

|
|||||
| Description | Fas-activated serine/threonine kinase (EC 2.7.11.8) (FAST kinase). | |||||
|
I17RB_HUMAN
|
||||||
| NC score | 0.013560 (rank : 17) | θ value | 5.27518 (rank : 16) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NRM6, Q9BPZ0, Q9NRL4, Q9NRM5 | Gene names | IL17RB, EVI27, IL17BR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-17 receptor B precursor (IL-17 receptor B) (IL-17RB) (Interleukin-17B receptor) (IL-17B receptor) (IL-17 receptor homolog 1) (IL-17Rh1) (IL17Rh1) (Cytokine receptor CRL4). | |||||
|
NUPL_HUMAN
|
||||||
| NC score | 0.012913 (rank : 18) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52594, Q15277, Q4VAS0, Q4VAS1, Q4VAS3, Q53QT8, Q53R11 | Gene names | HRB, RAB, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein) (Rev- interacting protein) (Rev/Rex activation domain-binding protein). | |||||
|
NUPL_MOUSE
|
||||||
| NC score | 0.012885 (rank : 19) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2K6, O70448, Q8BQL5, Q8CDK9 | Gene names | Hrb, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein homolog). | |||||
|
CP17A_HUMAN
|
||||||
| NC score | 0.007631 (rank : 20) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P05093, Q5TZV7 | Gene names | CYP17A1, CYP17, S17AH | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 17A1 (EC 1.14.99.9) (CYPXVII) (P450-C17) (P450c17) (Steroid 17-alpha-monooxygenase) (Steroid 17-alpha-hydroxylase/17,20 lyase). | |||||
|
SL9A2_HUMAN
|
||||||
| NC score | 0.005784 (rank : 21) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBY0 | Gene names | SLC9A2, NHE2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/hydrogen exchanger 2 (Na(+)/H(+) exchanger 2) (NHE-2) (Solute carrier family 9 member 2). | |||||
|
KCNB2_HUMAN
|
||||||
| NC score | 0.005644 (rank : 22) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.003875 (rank : 23) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.003812 (rank : 24) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.003274 (rank : 25) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
INADL_HUMAN
|
||||||
| NC score | 0.003198 (rank : 26) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
FOXC2_HUMAN
|
||||||
| NC score | 0.003167 (rank : 27) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99958 | Gene names | FOXC2, FKHL14, MFH1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C2 (Forkhead-related protein FKHL14) (Mesenchyme fork head protein 1) (MFH-1 protein) (Transcription factor FKH-14). | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.001719 (rank : 28) | θ value | 1.81305 (rank : 12) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.001131 (rank : 29) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
ZN287_HUMAN
|
||||||
| NC score | -0.001943 (rank : 30) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBT7 | Gene names | ZNF287 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 287. | |||||