Please be patient as the page loads
|
NUPL_MOUSE
|
||||||
| SwissProt Accessions | Q8K2K6, O70448, Q8BQL5, Q8CDK9 | Gene names | Hrb, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NUPL_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.985829 (rank : 2) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P52594, Q15277, Q4VAS0, Q4VAS1, Q4VAS3, Q53QT8, Q53R11 | Gene names | HRB, RAB, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein) (Rev- interacting protein) (Rev/Rex activation domain-binding protein). | |||||
|
NUPL_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q8K2K6, O70448, Q8BQL5, Q8CDK9 | Gene names | Hrb, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein homolog). | |||||
|
HRBL_MOUSE
|
||||||
| θ value | 5.17823e-64 (rank : 3) | NC score | 0.844370 (rank : 4) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
HRBL_HUMAN
|
||||||
| θ value | 8.55514e-59 (rank : 4) | NC score | 0.871035 (rank : 3) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
SMAP1_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 5) | NC score | 0.646065 (rank : 5) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
SMAP1_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 6) | NC score | 0.642116 (rank : 6) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
SMP1L_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 7) | NC score | 0.634269 (rank : 8) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WU79, Q5QPL2, Q96C93, Q9NST2, Q9UJL8 | Gene names | SMAP1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like. | |||||
|
SMP1L_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 8) | NC score | 0.634698 (rank : 7) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7TN29, Q3U798 | Gene names | Smap1l, Smap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like (Stromal membrane- associated protein 2). | |||||
|
CENA1_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 9) | NC score | 0.608823 (rank : 9) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75689 | Gene names | CENTA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 1 (Putative MAPK-activating protein PM25). | |||||
|
ARFG1_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 10) | NC score | 0.581676 (rank : 10) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N6T3, Q96KC4, Q96T02, Q9NSU3, Q9NVF6, Q9UIL0 | Gene names | ARFGAP1, ARF1GAP | |||
|
Domain Architecture |
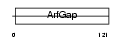
|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
CENG2_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 11) | NC score | 0.435932 (rank : 19) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BXK8, Q3UHA0, Q6ZPX9, Q8BZG0 | Gene names | Centg2, Agap1, Kiaa1099 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1). | |||||
|
CENG2_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 12) | NC score | 0.432675 (rank : 21) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UPQ3, Q541S5, Q6P9D7, Q9NV93 | Gene names | CENTG2, AGAP1, KIAA1099 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1) (GTP-binding and GTPase-activating protein 1) (GGAP1). | |||||
|
CENG3_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 13) | NC score | 0.439003 (rank : 18) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96P47, Q59EN0, Q96RK3 | Gene names | CENTG3, AGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
CENG3_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 14) | NC score | 0.439333 (rank : 16) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8VHH5, Q812F7 | Gene names | Centg3, Agap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
CENA2_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 15) | NC score | 0.572037 (rank : 11) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8R2V5 | Gene names | Centa2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 2. | |||||
|
ARFG1_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 16) | NC score | 0.569379 (rank : 12) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9EPJ9, Q3TI52, Q8BMM6, Q8BMQ7 | Gene names | Arfgap1, Arf1gap | |||
|
Domain Architecture |
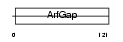
|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
CENB2_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 17) | NC score | 0.393167 (rank : 24) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
CENB5_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 18) | NC score | 0.400867 (rank : 22) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
CENG1_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 19) | NC score | 0.439087 (rank : 17) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
ARFG3_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 20) | NC score | 0.522131 (rank : 15) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9D8S3, Q99KN8 | Gene names | Arfgap3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
CENG1_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 21) | NC score | 0.433063 (rank : 20) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99490, O00578, Q548E0, Q8IWU3 | Gene names | CENTG1, AGAP2, KIAA0167 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE) (GTP-binding and GTPase-activating protein 2) (GGAP2). | |||||
|
CENA2_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 22) | NC score | 0.561111 (rank : 13) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NPF8, Q8N4Q6, Q96SD5 | Gene names | CENTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-alpha 2. | |||||
|
ARFG3_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 23) | NC score | 0.524538 (rank : 14) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NP61, Q9BSC6, Q9H9J0, Q9NT10, Q9NUP5, Q9Y4V3, Q9Y4V4 | Gene names | ARFGAP3, ARFGAP1 | |||
|
Domain Architecture |
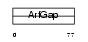
|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
CEND1_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 24) | NC score | 0.375498 (rank : 27) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
CEND3_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 25) | NC score | 0.369220 (rank : 31) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
CEND3_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 26) | NC score | 0.368250 (rank : 32) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
CEND1_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 27) | NC score | 0.371947 (rank : 30) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
DDEF1_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 28) | NC score | 0.383261 (rank : 26) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
DDEF2_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 29) | NC score | 0.373839 (rank : 28) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
CENB1_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 30) | NC score | 0.390671 (rank : 25) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
DDEF2_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 31) | NC score | 0.365873 (rank : 33) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
GIT2_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 32) | NC score | 0.316972 (rank : 35) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JLQ2 | Gene names | Git2 | |||
|
Domain Architecture |
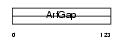
|
|||||
| Description | ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase- interactor 2) (Cool-interacting tyrosine-phosphorylated protein 2) (CAT2) (CAT-2). | |||||
|
CEND2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 33) | NC score | 0.354138 (rank : 34) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
DDFL1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 34) | NC score | 0.394571 (rank : 23) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8TDY4, Q6P9F4, Q86UY1, Q9NXK2 | Gene names | DDEFL1, UPLC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1 (Protein up- regulated in liver cancer 1). | |||||
|
DDFL1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 35) | NC score | 0.373596 (rank : 29) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5U464 | Gene names | Ddefl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1. | |||||
|
GP112_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 36) | NC score | 0.053057 (rank : 62) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 37) | NC score | 0.050997 (rank : 64) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
GIT2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 38) | NC score | 0.306432 (rank : 36) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14161, Q86U59, Q96CI2, Q9BV91, Q9Y5V2 | Gene names | GIT2, KIAA0148 | |||
|
Domain Architecture |
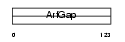
|
|||||
| Description | ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase- interactor 2) (GRK-interacting protein 2) (Cool-interacting tyrosine- phosphorylated protein 2) (CAT2) (CAT-2). | |||||
|
ADRM1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 39) | NC score | 0.072548 (rank : 40) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JKV1 | Gene names | Adrm1, Gp110 | |||
|
Domain Architecture |

|
|||||
| Description | Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110) (ARM-1). | |||||
|
GATA4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 40) | NC score | 0.027028 (rank : 82) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08369, P97491, Q9QZK4 | Gene names | Gata4, Gata-4 | |||
|
Domain Architecture |
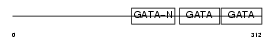
|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 0.21417 (rank : 41) | NC score | 0.046749 (rank : 68) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
CEP68_HUMAN
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.052069 (rank : 63) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q76N32, O60326, Q9BQ18, Q9UDM9 | Gene names | CEP68, KIAA0582 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
SYNG_HUMAN
|
||||||
| θ value | 0.279714 (rank : 43) | NC score | 0.060960 (rank : 51) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UMZ2, Q6ZT17 | Gene names | AP1GBP1, SYNG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
PER1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 44) | NC score | 0.036657 (rank : 73) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PPRB_MOUSE
|
||||||
| θ value | 0.365318 (rank : 45) | NC score | 0.048919 (rank : 66) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
SYNG_MOUSE
|
||||||
| θ value | 0.365318 (rank : 46) | NC score | 0.075717 (rank : 39) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
KLF4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 47) | NC score | 0.003284 (rank : 125) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 694 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60793, P70421 | Gene names | Klf4, Ezf, Gklf, Zie | |||
|
Domain Architecture |

|
|||||
| Description | Krueppel-like factor 4 (Gut-enriched krueppel-like factor) (Epithelial zinc-finger protein EZF). | |||||
|
PLK3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.000404 (rank : 129) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 865 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H4B4, Q15767, Q96CV1 | Gene names | PLK3, CNK, FNK, PRK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PLK3 (EC 2.7.11.21) (Polo-like kinase 3) (PLK-3) (Cytokine-inducible serine/threonine-protein kinase) (FGF- inducible kinase) (Proliferation-related kinase). | |||||
|
PO121_MOUSE
|
||||||
| θ value | 0.47712 (rank : 49) | NC score | 0.065662 (rank : 44) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.046442 (rank : 69) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.044187 (rank : 70) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.063632 (rank : 47) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
CD45_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.012529 (rank : 112) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P08575, Q16614, Q9H0Y6 | Gene names | PTPRC, CD45 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (CD45 antigen) (T200). | |||||
|
CNTN5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.005510 (rank : 122) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P68500 | Gene names | Cntn5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-5 precursor (Neural recognition molecule NB-2). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.021401 (rank : 87) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
ADRM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.061002 (rank : 50) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16186, Q9H1P2 | Gene names | ADRM1, GP110 | |||
|
Domain Architecture |
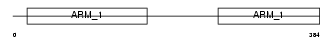
|
|||||
| Description | Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110). | |||||
|
EPN1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.034091 (rank : 76) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y6I3, Q86ST3, Q9HA18 | Gene names | EPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein). | |||||
|
PER1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.033470 (rank : 78) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.021583 (rank : 86) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.016856 (rank : 99) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.009520 (rank : 118) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
EP15_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.012659 (rank : 111) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.019236 (rank : 91) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
NU153_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.066863 (rank : 42) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
SNIP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.027532 (rank : 81) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9C0H9, Q8N4W8 | Gene names | P140, KIAA1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
CN032_MOUSE
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.063672 (rank : 46) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.021750 (rank : 85) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.017809 (rank : 96) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
PO121_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.071613 (rank : 41) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
TS1R2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.014308 (rank : 106) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q925I4, Q923J8 | Gene names | Tas1r2, Gpr71, T1r2, Tr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 2 precursor (G-protein coupled receptor 71) (Sweet taste receptor T1R2). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.026519 (rank : 83) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
EPN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.047298 (rank : 67) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
MISS_MOUSE
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.033529 (rank : 77) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
MNAB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.014646 (rank : 104) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P0C090 | Gene names | Mnab | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein. | |||||
|
RCOR1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.012256 (rank : 114) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UKL0, Q15044, Q6P2I9, Q86VG5 | Gene names | RCOR1, KIAA0071, RCOR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
SFRS8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.025061 (rank : 84) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12872 | Gene names | SFRS8, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 8 (Suppressor of white apricot protein homolog). | |||||
|
CK024_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.041927 (rank : 72) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
FLVC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.017393 (rank : 97) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5Y0, Q86XY9, Q9NVR9 | Gene names | FLVCR1, FLVCR | |||
|
Domain Architecture |

|
|||||
| Description | Feline leukemia virus subgroup C receptor-related protein 1 (Feline leukemia virus subgroup C receptor) (hFLVCR). | |||||
|
MCM3A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.018493 (rank : 94) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60318, Q9UMT4 | Gene names | MCM3AP, GANP, KIAA0572, MAP80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.027712 (rank : 80) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
T240L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.016805 (rank : 100) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q71F56, Q68DN4, Q9H8C0, Q9NSY9, Q9UFD8, Q9UPX5 | Gene names | THRAP2, KIAA1025, PROSIT240, TRAP240L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
TFR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.015524 (rank : 102) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62351, Q61560 | Gene names | Tfrc, Trfr | |||
|
Domain Architecture |

|
|||||
| Description | Transferrin receptor protein 1 (TfR1) (TR) (TfR) (Trfr) (CD71 antigen). | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.013916 (rank : 108) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
CN032_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.054675 (rank : 59) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
COHA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.017233 (rank : 98) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
FHOD1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.013236 (rank : 109) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P9Q4, Q8BMK2 | Gene names | Fhod1, Fhos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.016229 (rank : 101) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
HXD13_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.003372 (rank : 124) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35453 | Gene names | HOXD13, HOX4I | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D13 (Hox-4I). | |||||
|
KLF4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.001037 (rank : 128) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43474, P78338, Q9UNP3 | Gene names | KLF4, EZF, GKLF | |||
|
Domain Architecture |

|
|||||
| Description | Krueppel-like factor 4 (Epithelial zinc-finger protein EZF) (Gut- enriched krueppel-like factor). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.032501 (rank : 79) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PO2F1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.009297 (rank : 119) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P14859, Q5TBT7, Q6PK46, Q8NEU9, Q9BPV1 | Gene names | POU2F1, OCT1, OTF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 1 (Octamer-binding transcription factor 1) (Oct-1) (OTF-1) (NF-A1). | |||||
|
TECT1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.014311 (rank : 105) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BZ64, Q3UMP0, Q8BUE2 | Gene names | Tect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tectonic-1 precursor. | |||||
|
U689_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.021296 (rank : 88) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6UX39 | Gene names | ames=UNQ689/PRO1329 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein UNQ689/PRO1329 precursor. | |||||
|
ZN414_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.009689 (rank : 117) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 473 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DCK4, Q8CHU9 | Gene names | Znf414, Zfp414 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 414. | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.021099 (rank : 89) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
NUP62_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.042245 (rank : 71) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q63850, Q99JN7 | Gene names | Nup62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
P3H3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.012885 (rank : 110) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CG70, O88836 | Gene names | Leprel2, P3h3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 3 precursor (EC 1.14.11.7) (Leprecan-like protein 2) (Protein B). | |||||
|
PCD16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.001343 (rank : 127) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JQ0, O15098 | Gene names | DCHS1, CDH19, FIB1, KIAA1773, PCDH16 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-16 precursor (Dachsous 1) (Cadherin-19) (Fibroblast cadherin 1). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.007701 (rank : 121) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
UBP5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.019124 (rank : 93) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.019167 (rank : 92) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
BRWD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.002619 (rank : 126) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NSI6, O43721, Q5R2V0, Q5R2V1, Q8TCV3, Q96QG9, Q96QH0, Q9NUK1 | Gene names | BRWD1, C21orf107, WDR9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
CREM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.017888 (rank : 95) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03060, Q16114, Q16116, Q9NZB9 | Gene names | CREM | |||
|
Domain Architecture |
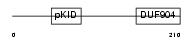
|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
DACH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.014060 (rank : 107) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UI36, O75523, O75687, Q5VYY3, Q5VYY4, Q96SG3, Q96SG4, Q9H524, Q9UMH4 | Gene names | DACH1, DACH | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.005505 (rank : 123) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
HTF4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.012017 (rank : 115) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99081 | Gene names | TCF12, HEB, HTF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4). | |||||
|
HTF4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.011529 (rank : 116) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61286 | Gene names | Tcf12, Alf1, Meb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4) (Class A helix-loop-helix transcription factor ME1). | |||||
|
LEUK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.034244 (rank : 75) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P16150 | Gene names | SPN, CD43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Galactoglycoprotein) (GALGP) (CD43 antigen). | |||||
|
LY75_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.012465 (rank : 113) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60767, Q8C7T3, Q91XL8, Q9QUZ6 | Gene names | Ly75, Cd205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (CD205 antigen). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.034877 (rank : 74) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.019926 (rank : 90) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PO2F1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.008323 (rank : 120) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P25425, Q61994, Q63891, Q6Y681, Q7TSD0, Q8BT04, Q8K570, Q99JH0, Q9WTZ4, Q9WTZ5, Q9WTZ6 | Gene names | Pou2f1, Oct-1, Otf-1, Otf1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 1 (Octamer-binding transcription factor 1) (Oct-1) (OTF-1) (NF-A1). | |||||
|
RD23B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.014849 (rank : 103) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
ANR46_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.050778 (rank : 65) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86W74, Q6P9B7 | Gene names | ANKRD46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 46 (Ankyrin repeat small protein) (ANK-S). | |||||
|
CO002_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.054694 (rank : 58) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
CT086_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.054627 (rank : 60) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BZ19, Q4VXE6 | Gene names | C20orf86 | |||
|
Domain Architecture |

|
|||||
| Description | Putative uncharacterized protein C20orf86. | |||||
|
DP13A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.056070 (rank : 54) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UKG1, Q9P2B9 | Gene names | APPL1, APPL, DIP13A, KIAA1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
DP13A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.055938 (rank : 55) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
DP13B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.053830 (rank : 61) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K3G9, Q99LT7 | Gene names | Dip13b, Appl2, Dip3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 beta (Dip13 beta) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 2). | |||||
|
ELN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.061132 (rank : 48) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P15502, O15336, O15337, Q14233, Q14234, Q14235, Q14238, Q6P0L4, Q6ZWJ6, Q75MU5, Q7Z316, Q7Z3F5, Q9UMF5 | Gene names | ELN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elastin precursor (Tropoelastin). | |||||
|
ELN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.063740 (rank : 45) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54320 | Gene names | Eln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elastin precursor (Tropoelastin). | |||||
|
GIT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.270554 (rank : 37) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y2X7, Q86SS0, Q9BRJ4 | Gene names | GIT1 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase- interactor 1) (GRK-interacting protein 1) (Cool-associated and tyrosine-phosphorylated protein 1) (Cat-1). | |||||
|
GIT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.270224 (rank : 38) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q68FF6 | Gene names | Git1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase- interactor 1) (GRK-interacting protein 1). | |||||
|
OPHN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.061067 (rank : 49) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
OPHN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.065805 (rank : 43) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
PKHA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.055710 (rank : 56) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.054826 (rank : 57) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PYGO2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.057229 (rank : 53) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BRQ0, Q8WYZ4, Q96CY2 | Gene names | PYGO2 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 2. | |||||
|
RHG26_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.057313 (rank : 52) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
NUPL_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q8K2K6, O70448, Q8BQL5, Q8CDK9 | Gene names | Hrb, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein homolog). | |||||
|
NUPL_HUMAN
|
||||||
| NC score | 0.985829 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P52594, Q15277, Q4VAS0, Q4VAS1, Q4VAS3, Q53QT8, Q53R11 | Gene names | HRB, RAB, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein) (Rev- interacting protein) (Rev/Rex activation domain-binding protein). | |||||
|
HRBL_HUMAN
|
||||||
| NC score | 0.871035 (rank : 3) | θ value | 8.55514e-59 (rank : 4) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
HRBL_MOUSE
|
||||||
| NC score | 0.844370 (rank : 4) | θ value | 5.17823e-64 (rank : 3) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
SMAP1_HUMAN
|
||||||
| NC score | 0.646065 (rank : 5) | θ value | 3.52202e-12 (rank : 5) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
SMAP1_MOUSE
|
||||||
| NC score | 0.642116 (rank : 6) | θ value | 3.52202e-12 (rank : 6) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
SMP1L_MOUSE
|
||||||
| NC score | 0.634698 (rank : 7) | θ value | 3.89403e-11 (rank : 8) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7TN29, Q3U798 | Gene names | Smap1l, Smap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like (Stromal membrane- associated protein 2). | |||||
|
SMP1L_HUMAN
|
||||||
| NC score | 0.634269 (rank : 8) | θ value | 3.89403e-11 (rank : 7) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WU79, Q5QPL2, Q96C93, Q9NST2, Q9UJL8 | Gene names | SMAP1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like. | |||||
|
CENA1_HUMAN
|
||||||
| NC score | 0.608823 (rank : 9) | θ value | 1.133e-10 (rank : 9) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75689 | Gene names | CENTA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 1 (Putative MAPK-activating protein PM25). | |||||
|
ARFG1_HUMAN
|
||||||
| NC score | 0.581676 (rank : 10) | θ value | 1.25267e-09 (rank : 10) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N6T3, Q96KC4, Q96T02, Q9NSU3, Q9NVF6, Q9UIL0 | Gene names | ARFGAP1, ARF1GAP | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
CENA2_MOUSE
|
||||||
| NC score | 0.572037 (rank : 11) | θ value | 1.69304e-06 (rank : 15) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8R2V5 | Gene names | Centa2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 2. | |||||
|
ARFG1_MOUSE
|
||||||
| NC score | 0.569379 (rank : 12) | θ value | 2.21117e-06 (rank : 16) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9EPJ9, Q3TI52, Q8BMM6, Q8BMQ7 | Gene names | Arfgap1, Arf1gap | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
CENA2_HUMAN
|
||||||
| NC score | 0.561111 (rank : 13) | θ value | 8.40245e-06 (rank : 22) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NPF8, Q8N4Q6, Q96SD5 | Gene names | CENTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-alpha 2. | |||||
|
ARFG3_HUMAN
|
||||||
| NC score | 0.524538 (rank : 14) | θ value | 4.1701e-05 (rank : 23) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NP61, Q9BSC6, Q9H9J0, Q9NT10, Q9NUP5, Q9Y4V3, Q9Y4V4 | Gene names | ARFGAP3, ARFGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
ARFG3_MOUSE
|
||||||
| NC score | 0.522131 (rank : 15) | θ value | 4.92598e-06 (rank : 20) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9D8S3, Q99KN8 | Gene names | Arfgap3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
CENG3_MOUSE
|
||||||
| NC score | 0.439333 (rank : 16) | θ value | 2.61198e-07 (rank : 14) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8VHH5, Q812F7 | Gene names | Centg3, Agap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
CENG1_MOUSE
|
||||||
| NC score | 0.439087 (rank : 17) | θ value | 3.77169e-06 (rank : 19) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
CENG3_HUMAN
|
||||||
| NC score | 0.439003 (rank : 18) | θ value | 2.61198e-07 (rank : 13) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96P47, Q59EN0, Q96RK3 | Gene names | CENTG3, AGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
CENG2_MOUSE
|
||||||
| NC score | 0.435932 (rank : 19) | θ value | 4.76016e-09 (rank : 11) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BXK8, Q3UHA0, Q6ZPX9, Q8BZG0 | Gene names | Centg2, Agap1, Kiaa1099 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1). | |||||
|
CENG1_HUMAN
|
||||||
| NC score | 0.433063 (rank : 20) | θ value | 4.92598e-06 (rank : 21) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99490, O00578, Q548E0, Q8IWU3 | Gene names | CENTG1, AGAP2, KIAA0167 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE) (GTP-binding and GTPase-activating protein 2) (GGAP2). | |||||
|
CENG2_HUMAN
|
||||||
| NC score | 0.432675 (rank : 21) | θ value | 1.38499e-08 (rank : 12) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UPQ3, Q541S5, Q6P9D7, Q9NV93 | Gene names | CENTG2, AGAP1, KIAA1099 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1) (GTP-binding and GTPase-activating protein 1) (GGAP1). | |||||
|
CENB5_HUMAN
|
||||||
| NC score | 0.400867 (rank : 22) | θ value | 3.77169e-06 (rank : 18) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
DDFL1_HUMAN
|
||||||
| NC score | 0.394571 (rank : 23) | θ value | 0.0113563 (rank : 34) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8TDY4, Q6P9F4, Q86UY1, Q9NXK2 | Gene names | DDEFL1, UPLC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1 (Protein up- regulated in liver cancer 1). | |||||
|
CENB2_HUMAN
|
||||||
| NC score | 0.393167 (rank : 24) | θ value | 2.21117e-06 (rank : 17) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
CENB1_HUMAN
|
||||||
| NC score | 0.390671 (rank : 25) | θ value | 0.00175202 (rank : 30) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
DDEF1_MOUSE
|
||||||
| NC score | 0.383261 (rank : 26) | θ value | 0.000786445 (rank : 28) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
CEND1_MOUSE
|
||||||
| NC score | 0.375498 (rank : 27) | θ value | 4.1701e-05 (rank : 24) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
DDEF2_HUMAN
|
||||||
| NC score | 0.373839 (rank : 28) | θ value | 0.00134147 (rank : 29) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
DDFL1_MOUSE
|
||||||
| NC score | 0.373596 (rank : 29) | θ value | 0.0148317 (rank : 35) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5U464 | Gene names | Ddefl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1. | |||||
|
CEND1_HUMAN
|
||||||
| NC score | 0.371947 (rank : 30) | θ value | 0.00020696 (rank : 27) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
CEND3_MOUSE
|
||||||
| NC score | 0.369220 (rank : 31) | θ value | 9.29e-05 (rank : 25) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
CEND3_HUMAN
|
||||||
| NC score | 0.368250 (rank : 32) | θ value | 0.000121331 (rank : 26) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
DDEF2_MOUSE
|
||||||
| NC score | 0.365873 (rank : 33) | θ value | 0.00228821 (rank : 31) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
CEND2_HUMAN
|
||||||
| NC score | 0.354138 (rank : 34) | θ value | 0.0113563 (rank : 33) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
GIT2_MOUSE
|
||||||
| NC score | 0.316972 (rank : 35) | θ value | 0.00509761 (rank : 32) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JLQ2 | Gene names | Git2 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase- interactor 2) (Cool-interacting tyrosine-phosphorylated protein 2) (CAT2) (CAT-2). | |||||
|
GIT2_HUMAN
|
||||||
| NC score | 0.306432 (rank : 36) | θ value | 0.125558 (rank : 38) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14161, Q86U59, Q96CI2, Q9BV91, Q9Y5V2 | Gene names | GIT2, KIAA0148 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase- interactor 2) (GRK-interacting protein 2) (Cool-interacting tyrosine- phosphorylated protein 2) (CAT2) (CAT-2). | |||||
|
GIT1_HUMAN
|
||||||
| NC score | 0.270554 (rank : 37) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y2X7, Q86SS0, Q9BRJ4 | Gene names | GIT1 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase- interactor 1) (GRK-interacting protein 1) (Cool-associated and tyrosine-phosphorylated protein 1) (Cat-1). | |||||
|
GIT1_MOUSE
|
||||||
| NC score | 0.270224 (rank : 38) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q68FF6 | Gene names | Git1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase- interactor 1) (GRK-interacting protein 1). | |||||
|
SYNG_MOUSE
|
||||||
| NC score | 0.075717 (rank : 39) | θ value | 0.365318 (rank : 46) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
ADRM1_MOUSE
|
||||||
| NC score | 0.072548 (rank : 40) | θ value | 0.163984 (rank : 39) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JKV1 | Gene names | Adrm1, Gp110 | |||
|
Domain Architecture |

|
|||||
| Description | Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110) (ARM-1). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.071613 (rank : 41) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.066863 (rank : 42) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
OPHN1_MOUSE
|
||||||
| NC score | 0.065805 (rank : 43) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.065662 (rank : 44) | θ value | 0.47712 (rank : 49) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
ELN_MOUSE
|
||||||
| NC score | 0.063740 (rank : 45) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54320 | Gene names | Eln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elastin precursor (Tropoelastin). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.063672 (rank : 46) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.063632 (rank : 47) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
ELN_HUMAN
|
||||||
| NC score | 0.061132 (rank : 48) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P15502, O15336, O15337, Q14233, Q14234, Q14235, Q14238, Q6P0L4, Q6ZWJ6, Q75MU5, Q7Z316, Q7Z3F5, Q9UMF5 | Gene names | ELN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elastin precursor (Tropoelastin). | |||||
|
OPHN1_HUMAN
|
||||||
| NC score | 0.061067 (rank : 49) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
ADRM1_HUMAN
|
||||||
| NC score | 0.061002 (rank : 50) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16186, Q9H1P2 | Gene names | ADRM1, GP110 | |||
|
Domain Architecture |

|
|||||
| Description | Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110). | |||||
|
SYNG_HUMAN
|
||||||
| NC score | 0.060960 (rank : 51) | θ value | 0.279714 (rank : 43) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UMZ2, Q6ZT17 | Gene names | AP1GBP1, SYNG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
RHG26_HUMAN
|
||||||
| NC score | 0.057313 (rank : 52) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
PYGO2_HUMAN
|
||||||
| NC score | 0.057229 (rank : 53) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BRQ0, Q8WYZ4, Q96CY2 | Gene names | PYGO2 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 2. | |||||
|
DP13A_HUMAN
|
||||||
| NC score | 0.056070 (rank : 54) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UKG1, Q9P2B9 | Gene names | APPL1, APPL, DIP13A, KIAA1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
DP13A_MOUSE
|
||||||
| NC score | 0.055938 (rank : 55) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
PKHA1_HUMAN
|
||||||
| NC score | 0.055710 (rank : 56) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA1_MOUSE
|
||||||
| NC score | 0.054826 (rank : 57) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
CO002_HUMAN
|
||||||
| NC score | 0.054694 (rank : 58) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.054675 (rank : 59) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CT086_HUMAN
|
||||||
| NC score | 0.054627 (rank : 60) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BZ19, Q4VXE6 | Gene names | C20orf86 | |||
|
Domain Architecture |

|
|||||
| Description | Putative uncharacterized protein C20orf86. | |||||
|
DP13B_MOUSE
|
||||||
| NC score | 0.053830 (rank : 61) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K3G9, Q99LT7 | Gene names | Dip13b, Appl2, Dip3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 beta (Dip13 beta) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 2). | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.053057 (rank : 62) | θ value | 0.0431538 (rank : 36) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
CEP68_HUMAN
|
||||||
| NC score | 0.052069 (rank : 63) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q76N32, O60326, Q9BQ18, Q9UDM9 | Gene names | CEP68, KIAA0582 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.050997 (rank : 64) | θ value | 0.0431538 (rank : 37) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
ANR46_HUMAN
|
||||||
| NC score | 0.050778 (rank : 65) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86W74, Q6P9B7 | Gene names | ANKRD46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 46 (Ankyrin repeat small protein) (ANK-S). | |||||
|
PPRB_MOUSE
|
||||||
| NC score | 0.048919 (rank : 66) | θ value | 0.365318 (rank : 45) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
EPN1_MOUSE
|
||||||
| NC score | 0.047298 (rank : 67) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.046749 (rank : 68) | θ value | 0.21417 (rank : 41) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.046442 (rank : 69) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.044187 (rank : 70) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
NUP62_MOUSE
|
||||||
| NC score | 0.042245 (rank : 71) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q63850, Q99JN7 | Gene names | Nup62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.041927 (rank : 72) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
PER1_HUMAN
|
||||||
| NC score | 0.036657 (rank : 73) | θ value | 0.365318 (rank : 44) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.034877 (rank : 74) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
LEUK_HUMAN
|
||||||
| NC score | 0.034244 (rank : 75) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P16150 | Gene names | SPN, CD43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Galactoglycoprotein) (GALGP) (CD43 antigen). | |||||
|
EPN1_HUMAN
|
||||||
| NC score | 0.034091 (rank : 76) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y6I3, Q86ST3, Q9HA18 | Gene names | EPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.033529 (rank : 77) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
PER1_MOUSE
|
||||||
| NC score | 0.033470 (rank : 78) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.032501 (rank : 79) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.027712 (rank : 80) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
SNIP_HUMAN
|
||||||
| NC score | 0.027532 (rank : 81) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9C0H9, Q8N4W8 | Gene names | P140, KIAA1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
GATA4_MOUSE
|
||||||
| NC score | 0.027028 (rank : 82) | θ value | 0.163984 (rank : 40) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08369, P97491, Q9QZK4 | Gene names | Gata4, Gata-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.026519 (rank : 83) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
SFRS8_HUMAN
|
||||||
| NC score | 0.025061 (rank : 84) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12872 | Gene names | SFRS8, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 8 (Suppressor of white apricot protein homolog). | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.021750 (rank : 85) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.021583 (rank : 86) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.021401 (rank : 87) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
U689_HUMAN
|
||||||
| NC score | 0.021296 (rank : 88) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6UX39 | Gene names | ames=UNQ689/PRO1329 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein UNQ689/PRO1329 precursor. | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.021099 (rank : 89) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.019926 (rank : 90) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.019236 (rank : 91) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
UBP5_MOUSE
|
||||||
| NC score | 0.019167 (rank : 92) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP5_HUMAN
|
||||||
| NC score | 0.019124 (rank : 93) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
MCM3A_HUMAN
|
||||||
| NC score | 0.018493 (rank : 94) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60318, Q9UMT4 | Gene names | MCM3AP, GANP, KIAA0572, MAP80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
CREM_HUMAN
|
||||||
| NC score | 0.017888 (rank : 95) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03060, Q16114, Q16116, Q9NZB9 | Gene names | CREM | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.017809 (rank : 96) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
FLVC1_HUMAN
|
||||||
| NC score | 0.017393 (rank : 97) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5Y0, Q86XY9, Q9NVR9 | Gene names | FLVCR1, FLVCR | |||
|
Domain Architecture |

|
|||||
| Description | Feline leukemia virus subgroup C receptor-related protein 1 (Feline leukemia virus subgroup C receptor) (hFLVCR). | |||||
|
COHA1_MOUSE
|
||||||
| NC score | 0.017233 (rank : 98) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.016856 (rank : 99) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
T240L_HUMAN
|
||||||
| NC score | 0.016805 (rank : 100) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q71F56, Q68DN4, Q9H8C0, Q9NSY9, Q9UFD8, Q9UPX5 | Gene names | THRAP2, KIAA1025, PROSIT240, TRAP240L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.016229 (rank : 101) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
TFR1_MOUSE
|
||||||
| NC score | 0.015524 (rank : 102) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62351, Q61560 | Gene names | Tfrc, Trfr | |||
|
Domain Architecture |

|
|||||
| Description | Transferrin receptor protein 1 (TfR1) (TR) (TfR) (Trfr) (CD71 antigen). | |||||
|
RD23B_MOUSE
|
||||||
| NC score | 0.014849 (rank : 103) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
MNAB_MOUSE
|
||||||
| NC score | 0.014646 (rank : 104) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P0C090 | Gene names | Mnab | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein. | |||||
|
TECT1_MOUSE
|
||||||
| NC score | 0.014311 (rank : 105) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BZ64, Q3UMP0, Q8BUE2 | Gene names | Tect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tectonic-1 precursor. | |||||
|
TS1R2_MOUSE
|
||||||
| NC score | 0.014308 (rank : 106) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q925I4, Q923J8 | Gene names | Tas1r2, Gpr71, T1r2, Tr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 2 precursor (G-protein coupled receptor 71) (Sweet taste receptor T1R2). | |||||
|
DACH1_HUMAN
|
||||||
| NC score | 0.014060 (rank : 107) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UI36, O75523, O75687, Q5VYY3, Q5VYY4, Q96SG3, Q96SG4, Q9H524, Q9UMH4 | Gene names | DACH1, DACH | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.013916 (rank : 108) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
FHOD1_MOUSE
|
||||||
| NC score | 0.013236 (rank : 109) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P9Q4, Q8BMK2 | Gene names | Fhod1, Fhos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
P3H3_MOUSE
|
||||||
| NC score | 0.012885 (rank : 110) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CG70, O88836 | Gene names | Leprel2, P3h3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 3 precursor (EC 1.14.11.7) (Leprecan-like protein 2) (Protein B). | |||||
|
EP15_HUMAN
|
||||||
| NC score | 0.012659 (rank : 111) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
CD45_HUMAN
|
||||||
| NC score | 0.012529 (rank : 112) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P08575, Q16614, Q9H0Y6 | Gene names | PTPRC, CD45 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (CD45 antigen) (T200). | |||||
|
LY75_MOUSE
|
||||||
| NC score | 0.012465 (rank : 113) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60767, Q8C7T3, Q91XL8, Q9QUZ6 | Gene names | Ly75, Cd205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (CD205 antigen). | |||||
|
RCOR1_HUMAN
|
||||||
| NC score | 0.012256 (rank : 114) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UKL0, Q15044, Q6P2I9, Q86VG5 | Gene names | RCOR1, KIAA0071, RCOR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
HTF4_HUMAN
|
||||||
| NC score | 0.012017 (rank : 115) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99081 | Gene names | TCF12, HEB, HTF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4). | |||||
|
HTF4_MOUSE
|
||||||
| NC score | 0.011529 (rank : 116) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61286 | Gene names | Tcf12, Alf1, Meb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4) (Class A helix-loop-helix transcription factor ME1). | |||||
|
ZN414_MOUSE
|
||||||
| NC score | 0.009689 (rank : 117) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 473 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DCK4, Q8CHU9 | Gene names | Znf414, Zfp414 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 414. | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.009520 (rank : 118) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
PO2F1_HUMAN
|
||||||
| NC score | 0.009297 (rank : 119) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P14859, Q5TBT7, Q6PK46, Q8NEU9, Q9BPV1 | Gene names | POU2F1, OCT1, OTF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 1 (Octamer-binding transcription factor 1) (Oct-1) (OTF-1) (NF-A1). | |||||
|
PO2F1_MOUSE
|
||||||
| NC score | 0.008323 (rank : 120) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P25425, Q61994, Q63891, Q6Y681, Q7TSD0, Q8BT04, Q8K570, Q99JH0, Q9WTZ4, Q9WTZ5, Q9WTZ6 | Gene names | Pou2f1, Oct-1, Otf-1, Otf1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 1 (Octamer-binding transcription factor 1) (Oct-1) (OTF-1) (NF-A1). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.007701 (rank : 121) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
CNTN5_MOUSE
|
||||||
| NC score | 0.005510 (rank : 122) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P68500 | Gene names | Cntn5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-5 precursor (Neural recognition molecule NB-2). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.005505 (rank : 123) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
HXD13_HUMAN
|
||||||
| NC score | 0.003372 (rank : 124) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35453 | Gene names | HOXD13, HOX4I | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D13 (Hox-4I). | |||||
|
KLF4_MOUSE
|
||||||
| NC score | 0.003284 (rank : 125) | θ value | 0.47712 (rank : 47) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 694 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60793, P70421 | Gene names | Klf4, Ezf, Gklf, Zie | |||
|
Domain Architecture |

|
|||||
| Description | Krueppel-like factor 4 (Gut-enriched krueppel-like factor) (Epithelial zinc-finger protein EZF). | |||||
|
BRWD1_HUMAN
|
||||||
| NC score | 0.002619 (rank : 126) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NSI6, O43721, Q5R2V0, Q5R2V1, Q8TCV3, Q96QG9, Q96QH0, Q9NUK1 | Gene names | BRWD1, C21orf107, WDR9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
PCD16_HUMAN
|
||||||
| NC score | 0.001343 (rank : 127) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JQ0, O15098 | Gene names | DCHS1, CDH19, FIB1, KIAA1773, PCDH16 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-16 precursor (Dachsous 1) (Cadherin-19) (Fibroblast cadherin 1). | |||||
|
KLF4_HUMAN
|
||||||
| NC score | 0.001037 (rank : 128) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43474, P78338, Q9UNP3 | Gene names | KLF4, EZF, GKLF | |||
|
Domain Architecture |

|
|||||
| Description | Krueppel-like factor 4 (Epithelial zinc-finger protein EZF) (Gut- enriched krueppel-like factor). | |||||
|
PLK3_HUMAN
|
||||||
| NC score | 0.000404 (rank : 129) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 865 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H4B4, Q15767, Q96CV1 | Gene names | PLK3, CNK, FNK, PRK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PLK3 (EC 2.7.11.21) (Polo-like kinase 3) (PLK-3) (Cytokine-inducible serine/threonine-protein kinase) (FGF- inducible kinase) (Proliferation-related kinase). | |||||